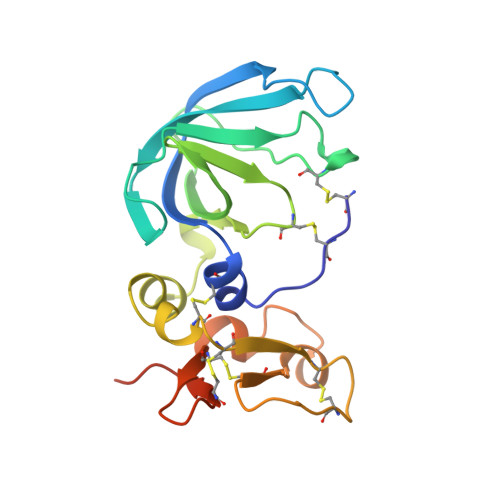
Three-dimensional structure of human tissue inhibitor of metalloproteinases-2 at 2.1 A resolution.
Tuuttila, A., Morgunova, E., Bergmann, U., Lindqvist, Y., Maskos, K., Fernandez-Catalan, C., Bode, W., Tryggvason, K., Schneider, G.(1998) J Mol Biology 284: 1133-1140
- PubMed: 9837731
- DOI: https://doi.org/10.1006/jmbi.1998.2223
- Primary Citation of Related Structures:
1BR9 - PubMed Abstract:
The three-dimensional structure of human tissue inhibitor of metalloproteinases-2 (TIMP-2) was determined by X-ray crystallography to 2.1 A resolution. The structure of the inhibitor consists of two domains. The N-terminal domain (residues 1-110) is folded into a beta-barrel, similar to the oligonucleotide/oligosaccharide binding fold otherwise found in certain DNA-binding proteins. The C-terminal domain (residues 111-194) contains a parallel stranded beta-hairpin plus a beta-loop-beta motif. Comparison of the structure of uncomplexed human TIMP-2 with that of bovine TIMP-2 bound to the catalytic domain of human MMP-14 suggests an internal rotation between the two domains of approximately 13 degrees upon binding to the protease. Furthermore, local conformational differences in the two structures that might be induced by formation of the protease-inhibitor complex have been found. The most prominent of these involves residues 27-40 of the A-B beta-hairpin loop. Structure-based alignment of amino acid sequences of representatives of the TIMP family maps the sequence differences mainly to loop regions, and some of these differences are proposed to be responsible for the particular properties of the various TIMP species.
- Department of Medical Biochemistry and Biophysics Division of Matrix Biology, Karolinska Institutet, Stockholm, S-171 77, Sweden.
Organizational Affiliation: