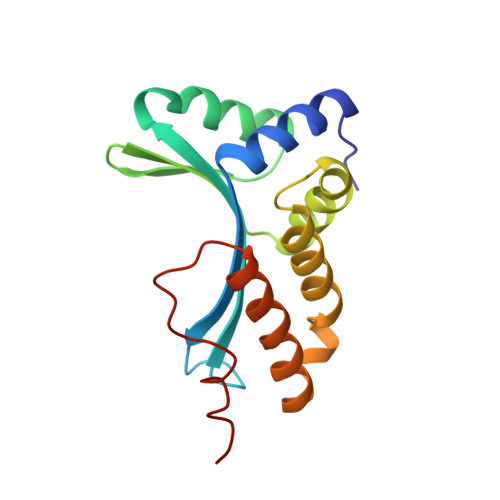
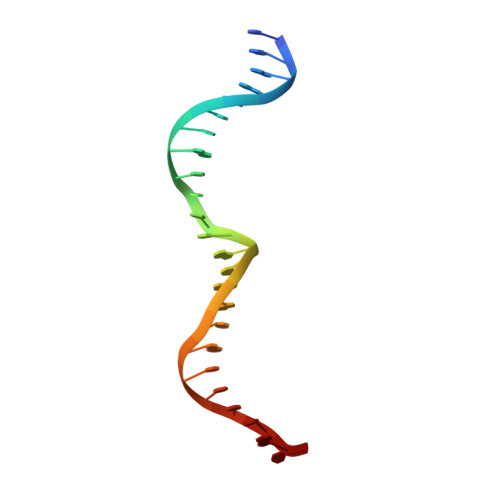
DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-CreI.
Jurica, M.S., Monnat Jr., R.J., Stoddard, B.L.(1998) Mol Cell 2: 469-476
- PubMed: 9809068
- DOI: https://doi.org/10.1016/s1097-2765(00)80146-x
- Primary Citation of Related Structures:
1BP7 - PubMed Abstract:
The structure of the LAGLIDADG intron-encoded homing endonuclease I-CreI bound to homing site DNA has been determined. The interface is formed by an extended, concave beta sheet from each enzyme monomer that contacts each DNA half-site, resulting in direct side-chain contacts to 18 of the 24 base pairs across the full-length homing site. The structure indicates that I-CreI is optimized to its role in genetic transposition by exhibiting long site-recognition while being able to cleave many closely related target sequences. DNA cleavage is mediated by a compact pair of active sites in the I-CreI homodimer, each of which contains a separate bound divalent cation.
- Fred Hutchinson Cancer Research Center, University of Washington, Seattle, Washington 98109, USA.
Organizational Affiliation: