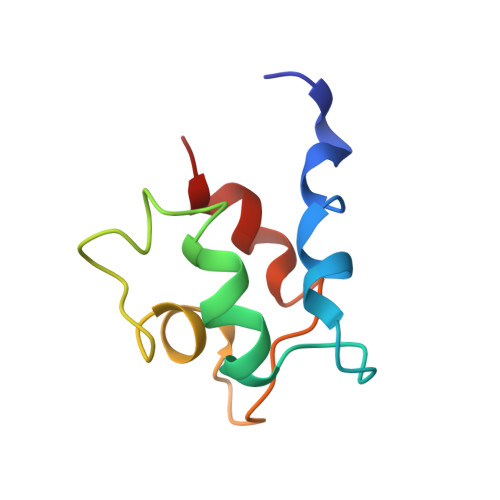
The solution structures of mutant calbindin D9k's, as determined by NMR, show that the calcium-binding site can adopt different folds.
Johansson, C., Ullner, M., Drakenberg, T.(1993) Biochemistry 32: 8429-8438
- PubMed: 8357794
- Primary Citation of Related Structures:
1BOC, 1BOD - PubMed Abstract:
The complete 1H NMR assignments have been obtained for five mutant proteins of calbindin D9k and the three-dimensional solution structures determined for two of the mutants. The structures have been determined using distance geometry and simulated annealing, with distance constraints from NMR. All mutants have modifications in the first calcium-binding site of calbindin (the N-terminal site designated the pseudo-EF-hand). The 3D structure of the mutant with the most extensive modifications in the pseudo-EF-hand shows that the site has turned inside-out and coordinates calcium as in the normal EF-hand (the C-terminal site). In a pseudo-EF-hand loop the calcium is coordinated by main-chain carbonyls, whereas calcium in the normal EF-hand is coordinated by side-chain carboxylates. The 3D structures and 1H NMR assignments show that in order to accomplish a change in the coordinating ligands of the pseudo-EF-hand the loop must be 12 residues long and have glycine in the sixth position. It does, however, seem possible to have alanine instead of aspartic acid in the first calcium coordinating position. The overall global fold of the proteins has not been affected by the mutations in the calcium-binding site, as compared to the wild-type calbindin D9k [Kördel, J., Skelton, N. J., Akke, M., & Chazin, W. J. (1993) J. Mol. Biol. (in press)]. The structures consist of two helix-calcium-binding loop-helix motifs, the so called EF-hands, and the loops are connected by a short antiparallel beta-sheet. All helices are pairwise in an antiparallel orientation.
- Chemical Center, Lund University, Sweden.
Organizational Affiliation: