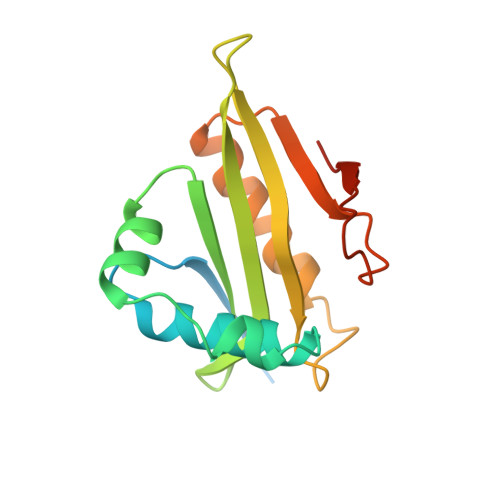
Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase.
Wolf, E., Vassilev, A., Makino, Y., Sali, A., Nakatani, Y., Burley, S.K.(1998) Cell 94: 439-449
- PubMed: 9727487
- DOI: https://doi.org/10.1016/s0092-8674(00)81585-8
- Primary Citation of Related Structures:
1BO4 - PubMed Abstract:
The X-ray structure of a canonical GCN5-related N-acetyltransferase (GNAT), Serratia marcescens aminoglycoside 3-N-acetyltransferase, bound to coenzyme A (CoA) has been determined at 2.3 A resolution. The single domain alpha/beta protein resembles a cupped right hand wrapped around a cylinder and consists of a highly curved, six-stranded beta sheet of mixed polarity that is sandwiched between four alpha helices. The structure includes all four conserved GNAT motifs (C, D, A, and B) and represents the catalytic core of this large enzyme superfamily. Acetyl CoA recognition is mediated by a betaalpha structure derived from GNAT motif A, which presents an invariant Arg/Gln-X-X-Gly-X-Gly/Ala segment for hydrogen bonding with the cofactor. Motif B contributes acidic residues to the binding site for the positively charged antibiotic substrate.
- Laboratories of Molecular Biophysics, The Rockefeller University, New York, New York 10021, USA.
Organizational Affiliation: