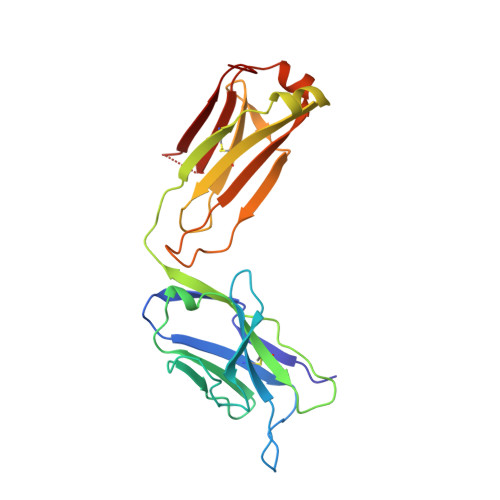
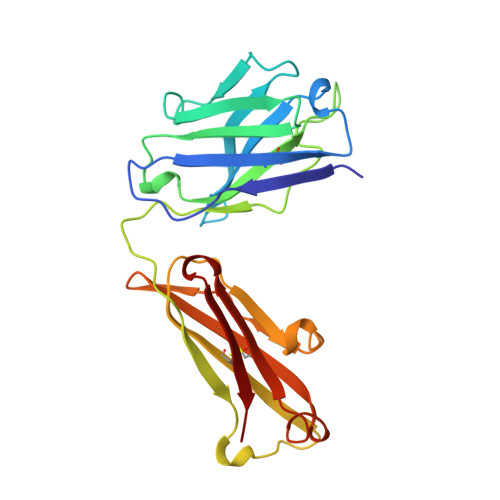
Mode of binding of anti-P-glycoprotein antibody MRK-16 to its antigen. A crystallographic and molecular modeling study.
Vasudevan, S., Tsuruo, T., Rose, D.R.(1998) J Biological Chem 273: 25413-25419
- PubMed: 9738009
- DOI: https://doi.org/10.1074/jbc.273.39.25413
- Primary Citation of Related Structures:
1BLN - PubMed Abstract:
Monoclonal antibody MRK-16 recognizes a discontinuous extracellular epitope on the multidrug resistance-associated ATP-binding cassette transporter, P-glycoprotein. The atomic basis for specificity of this antibody is of interest because of its potential as a modulator of P-glycoprotein activity. The crystal structure of Fab MRK-16 is reported to a resolution of 2.8 A. A structure for a portion of the epitope was derived by comparison to regions of solved structures with similar primary sequence. This has permitted a proposal for the mode of binding of the peptide epitope to the antibody, in which the peptide makes specific contacts with complementarity-determining regions H1, H2, and H3 from the heavy chain and L3 from the light chain. These interactions are consistent with epitope mapping studies and with the observation that MRK-16 is specific for human class I P-glycoprotein. This result identifies side chains in MRK-16 that would be amenable to alteration in antibody engineering experiments to derive improved multidrug resistance inhibitors for clinical use during chemotherapy. In particular, Arg-H97 contacts both Glu-746 and Asp-744 of the peptide, Arg-L96 contacts Asp-743, and Thr-H33 interacts with Thr-747. All of these epitope residues were implicated in mediating specificity by epitope mapping studies.
- Ontario Cancer Institute and Department of Medical Biophysics, University of Toronto, Toronto M5G 2M9, Canada.
Organizational Affiliation: