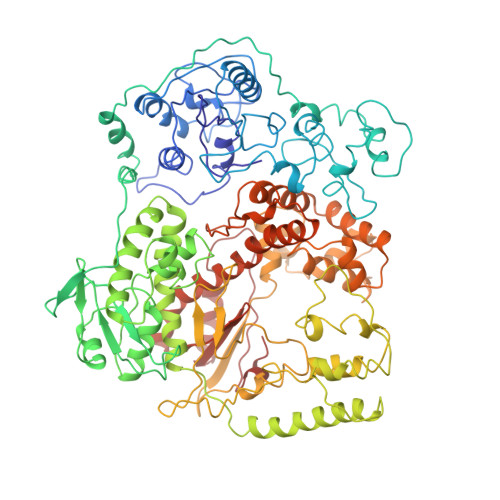
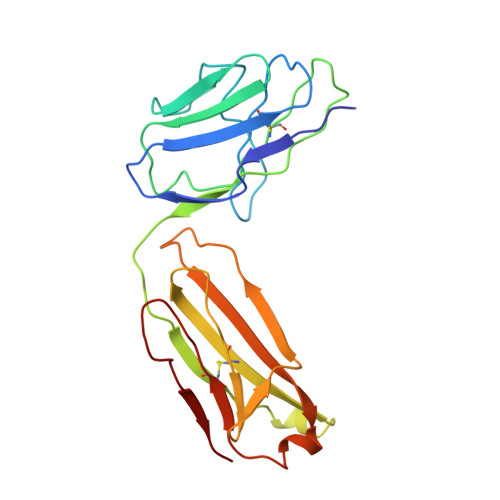
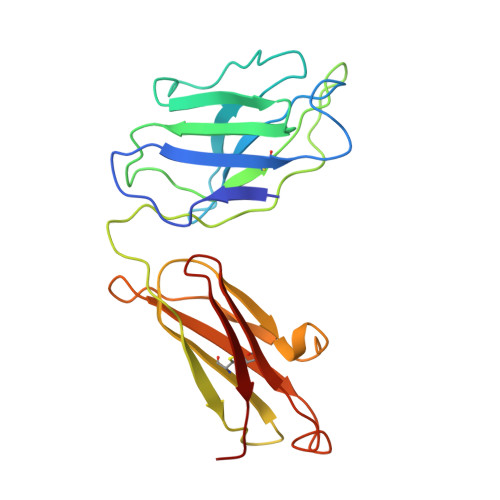
Crystal structure of Taq DNA polymerase in complex with an inhibitory Fab: the Fab is directed against an intermediate in the helix-coil dynamics of the enzyme.
Murali, R., Sharkey, D.J., Daiss, J.L., Murthy, H.M.(1998) Proc Natl Acad Sci U S A 95: 12562-12567
- PubMed: 9770525
- DOI: https://doi.org/10.1073/pnas.95.21.12562
- Primary Citation of Related Structures:
1BGX - PubMed Abstract:
We report the crystal structure of Thermus aquaticus DNA polymerase I in complex with an inhibitory Fab, TP7, directed against the native enzyme. Some of the residues present in a helical conformation in the native enzyme have adopted a gamma turn conformation in the complex. Taken together, structural information that describes alteration of helical structure and solution studies that demonstrate the ability of TP7 to inhibit 100% of the polymerase activity of the enzyme suggest that the change in conformation is probably caused by trapping of an intermediate in the helix-coil dynamics of this helix by the Fab. Antibodies directed against modified helices in proteins have long been anticipated. The present structure provides direct crystallographic evidence. The Fab binds within the DNA binding cleft of the polymerase domain, interacting with several residues that are used by the enzyme in binding the primer:template complex. This result unequivocally corroborates inferences drawn from binding experiments and modeling calculations that the inhibitory activity of this Fab is directly attributable to its interference with DNA binding by the polymerase domain of the enzyme. The combination of interactions made by the Fab residues in both the polymerase and the vestigial editing nuclease domain of the enzyme reveal the structural basis of its preference for binding to DNA polymerases of the Thermus species. The orientation of the structure-specific nuclease domain with respect to the polymerase domain is significantly different from that seen in other structures of this polymerase. This reorientation does not appear to be antibody-induced and implies remarkably high relative mobility between these two domains.
- Department of Pathology, University of Pennsylvania, Philadelphia, PA, 19104, USA.
Organizational Affiliation: