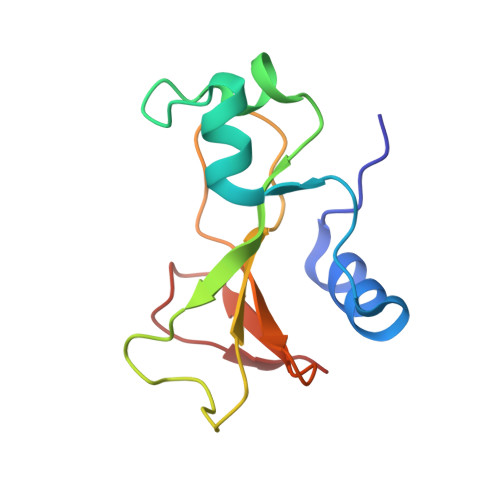
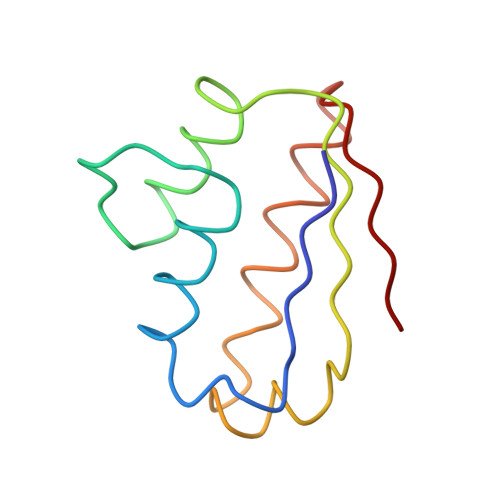
Recognition between a bacterial ribonuclease, barnase, and its natural inhibitor, barstar.
Guillet, V., Lapthorn, A., Hartley, R.W., Mauguen, Y.(1993) Structure 1: 165-176
- PubMed: 16100951
- DOI: https://doi.org/10.1016/0969-2126(93)90018-c
- Primary Citation of Related Structures:
1BGS - PubMed Abstract:
Protein-protein recognition is fundamental to most biological processes. The information we have so far on the interfaces between proteins comes largely from several protease-inhibitor and antigen-antibody complexes. Barnase, a bacterial ribonuclease, and barstar, its natural inhibitor, form a tight complex which provides a good model for the study and design of protein-protein non-covalent interactions. Here we report the structure of a complex between barnase and a fully functional mutant of barstar determined by X-ray analysis. Barstar is composed of three parallel alpha-helices stacked against a three-stranded parallel, beta-sheet, and sterically blocks the active site of the enzyme with an alpha-helix and adjacent loop. The buried surface in the interface between the two molecules totals 1630 A2. The barnase-barstar complex is predominantly stabilized by charge interactions involving positive charges in the active site of the enzyme. Asp39 of barstar binds to the phosphate-binding site of barnase, mimicking enzyme-substrate interactions. The phosphate-binding site of the enzyme is the anchor point for inhibitor binding. We propose that this is also likely to be the case for other ribonuclease inhibitors.
- Laboratoire de Physique, CNRS, UPR 180, Centre d'Etudes Pharmaceutiques, 92296 Châtenay-Malabry Cedex, France.
Organizational Affiliation: