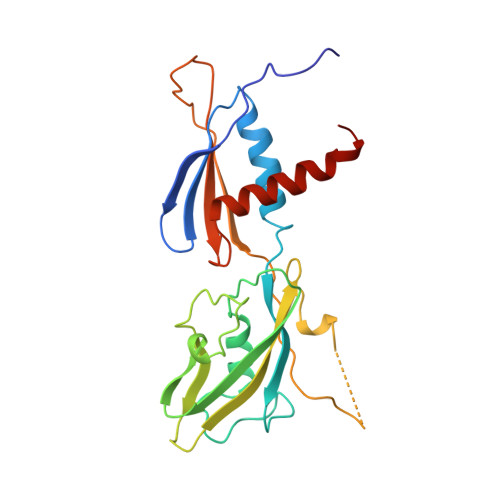
Structure of the Escherichia coli RNA polymerase alpha subunit amino-terminal domain.
Zhang, G., Darst, S.A.(1998) Science 281: 262-266
- PubMed: 9657722
- DOI: https://doi.org/10.1126/science.281.5374.262
- Primary Citation of Related Structures:
1BDF - PubMed Abstract:
The 2.5 angstrom resolution x-ray crystal structure of the Escherichia coli RNA polymerase (RNAP) alpha subunit amino-terminal domain (alphaNTD), which is necessary and sufficient to dimerize and assemble the other RNAP subunits into a transcriptionally active enzyme and contains all of the sequence elements conserved among eukaryotic alpha homologs, has been determined. The alphaNTD monomer comprises two distinct, flexibly linked domains, only one of which participates in the dimer interface. In the alphaNTD dimer, a pair of helices from one monomer interact with the cognate helices of the other to form an extensive hydrophobic core. All of the determinants for interactions with the other RNAP subunits lie on one face of the alphaNTD dimer. Sequence alignments, combined with secondary-structure predictions, support proposals that a heterodimer of the eukaryotic RNAP subunits related to Saccharomyces cerevisiae Rpb3 and Rpb11 plays the role of the alphaNTD dimer in prokaryotic RNAP.
- Rockefeller University, 1230 York Avenue, New York, NY 10021, USA.
Organizational Affiliation: