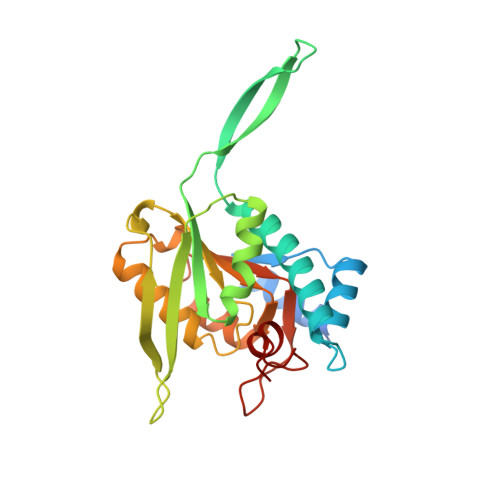
Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
Schumacher, M.A., Carter, D., Scott, D.M., Roos, D.S., Ullman, B., Brennan, R.G.(1998) EMBO J 17: 3219-3232
- PubMed: 9628859
- DOI: https://doi.org/10.1093/emboj/17.12.3219
- Primary Citation of Related Structures:
1BD3, 1BD4, 1UPF, 1UPU - PubMed Abstract:
Uracil phosphoribosyltransferase (UPRTase) catalyzes the transfer of a ribosyl phosphate group from alpha-D-5-phosphoribosyl-1-pyrophosphate to the N1 nitrogen of uracil. The UPRTase from the opportunistic pathogen Toxoplasma gondii is a rational target for antiparasitic drug design. To aid in structure-based drug design studies against toxoplasmosis, the crystal structures of the T.gondii apo UPRTase (1.93 A resolution), the UPRTase bound to its substrate, uracil (2.2 A resolution), its product, UMP (2.5 A resolution), and the prodrug, 5-fluorouracil (2.3 A resolution), have been determined. These structures reveal that UPRTase recognizes uracil through polypeptide backbone hydrogen bonds to the uracil exocyclic O2 and endocyclic N3 atoms and a backbone-water-exocyclic O4 oxygen hydrogen bond. This stereochemical arrangement and the architecture of the uracil-binding pocket reveal why cytosine and pyrimidines with exocyclic substituents at ring position 5 larger than fluorine, including thymine, cannot bind to the enzyme. Strikingly, the T. gondii UPRTase contains a 22 residue insertion within the conserved PRTase fold that forms an extended antiparallel beta-arm. Leu92, at the tip of this arm, functions to cap the active site of its dimer mate, thereby inhibiting the escape of the substrate-binding water molecule.
- Department of Biochemistry and Molecular Biology, Oregon Health Sciences University, Portland, OR 97201-3098, USA.
Organizational Affiliation: