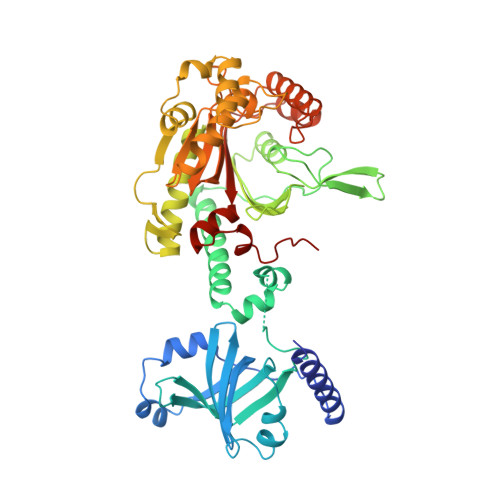
Structural studies of lysyl-tRNA synthetase: conformational changes induced by substrate binding.
Onesti, S., Desogus, G., Brevet, A., Chen, J., Plateau, P., Blanquet, S., Brick, P.(2000) Biochemistry 39: 12853-12861
- PubMed: 11041850
- DOI: https://doi.org/10.1021/bi001487r
- Primary Citation of Related Structures:
1BBU, 1BBW - PubMed Abstract:
Lysyl-tRNA synthetase is a member of the class II aminoacyl-tRNA synthetases and catalyses the specific aminoacylation of tRNA(Lys). The crystal structure of the constitutive lysyl-tRNA synthetase (LysS) from Escherichia coli has been determined to 2.7 A resolution in the unliganded form and in a complex with the lysine substrate. A comparison between the unliganded and lysine-bound structures reveals major conformational changes upon lysine binding. The lysine substrate is involved in a network of hydrogen bonds. Two of these interactions, one between the alpha-amino group and the carbonyl oxygen of Gly 216 and the other between the carboxylate group and the side chain of Arg 262, trigger a subtle and complicated reorganization of the active site, involving the ordering of two loops (residues 215-217 and 444-455), a change in conformation of residues 393-409, and a rotation of a 4-helix bundle domain (located between motif 2 and 3) by 10 degrees. The result of these changes is a closing up of the active site upon lysine binding.
- Biophysics Section, Blackett Laboratory, Imperial College, London SW7 2BZ, UK. s.onesti@ic.ac.uk
Organizational Affiliation: