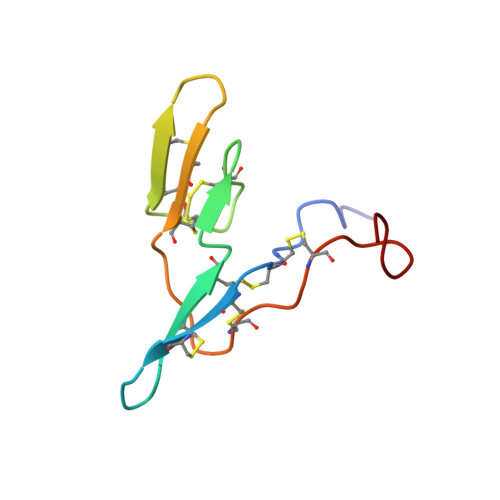
Three-dimensional structure of soybean trypsin/chymotrypsin Bowman-Birk inhibitor in solution.
Werner, M.H., Wemmer, D.E.(1992) Biochemistry 31: 999-1010
- PubMed: 1734975
- DOI: https://doi.org/10.1021/bi00119a008
- Primary Citation of Related Structures:
1BBI, 2BBI - PubMed Abstract:
The three-dimensional structure of soybean trypsin/chymotrypsin Bowman-Birk inhibitor in solution has been determined by two-dimensional 1H nuclear magnetic resonance spectroscopy and dynamical simulated annealing using the program XPLOR. The structure was defined by 907 NOEs involving intra- and interresidue contacts which served as distance constraints for a protocol of dynamical simulated annealing. In addition, 48 phi angle constraints involving non-proline amino acids, 29 chi angle constraints, six omega angle constraints for the X-Pro peptide bond, and 35 stereoassignments for prochiral centers were incorporated during the course of the calculation. The protein is characterized by two distinct binding domains for serine protease. Each domain is comprised of a beta-hairpin (antiparallel beta-sheet and a cis-proline-containing type VIb reverse turn) with a short segment making a third strand of antiparallel beta-sheet. The structure determination and refinement are described, and the structure is compared to other structures of Bowman-Birk inhibitors as well as other families of serine protease inhibitors.
- Chemical Biodynamics Division, Lawrence Berkeley Laboratory, University of California 94720.
Organizational Affiliation: