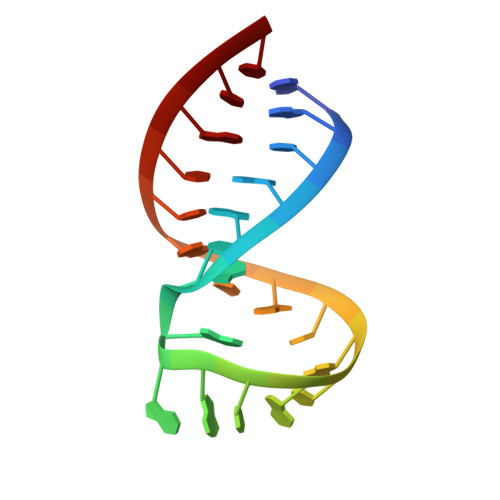
Structure of the dimer initiation complex of HIV-1 genomic RNA.
Mujeeb, A., Clever, J.L., Billeci, T.M., James, T.L., Parslow, T.G.(1998) Nat Struct Biol 5: 432-436
- PubMed: 9628479
- DOI: https://doi.org/10.1038/nsb0698-432
- Primary Citation of Related Structures:
1BAU - PubMed Abstract:
Retroviral genomes must dimerize to be fully infectious. Dimerization is directed by a unique RNA hairpin structure with a palindrome in its loop: hairpins of two strands first associate transiently through their loops, and then refold to a more stable, linear duplex. The structure of the initial, kissing-loop dimer from HIV-1, solved using 2D NMR, is bent and metastable, its interface being formed not only by standard basepairing between palindromes, but also by a distinctive pattern of interstrand stacking among bases at the stem-loop junctions. This creates mechanical distortions that partially melt both stems, which may facilitate spontaneous refolding of this RNA complex into linear form.
- Department of Pathology, University of California at San Francisco, 94143, USA.
Organizational Affiliation: