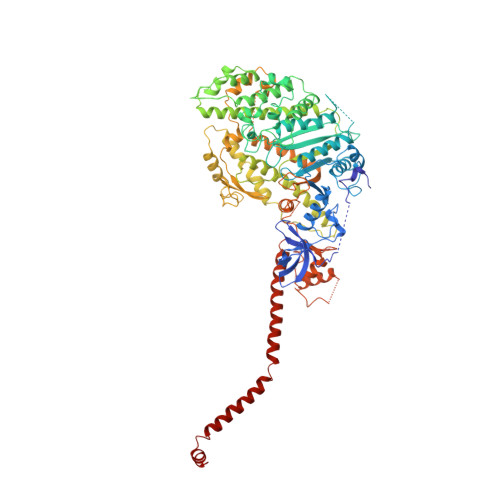
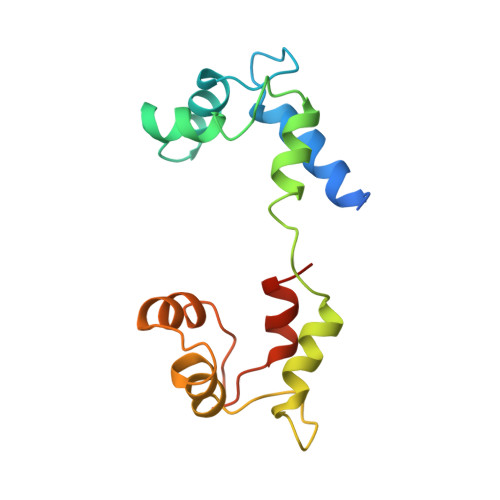
Atomic structure of scallop myosin subfragment S1 complexed with MgADP: a novel conformation of the myosin head.
Houdusse, A., Kalabokis, V.N., Himmel, D., Szent-Gyorgyi, A.G., Cohen, C.(1999) Cell 97: 459-470
- PubMed: 10338210
- DOI: https://doi.org/10.1016/s0092-8674(00)80756-4
- Primary Citation of Related Structures:
1B7T - PubMed Abstract:
The crystal structure of a proteolytic subfragment from scallop striated muscle myosin, complexed with MgADP, has been solved at 2.5 A resolution and reveals an unusual conformation of the myosin head. The converter and the lever arm are in very different positions from those in either the pre-power stroke or near-rigor state structures; moreover, in contrast to these structures, the SH1 helix is seen to be unwound. Here we compare the overall organization of the myosin head in these three states and show how the conformation of three flexible "joints" produces rearrangements of the four major subdomains in the myosin head with different bound nucleotides. We believe that this novel structure represents one of the prehydrolysis ("ATP") states of the contractile cycle in which the myosin heads stay detached from actin.
- Rosenstiel Basic Medical Sciences Research Center, Brandeis University, Waltham, Massachusetts 02254-9110, USA.
Organizational Affiliation: