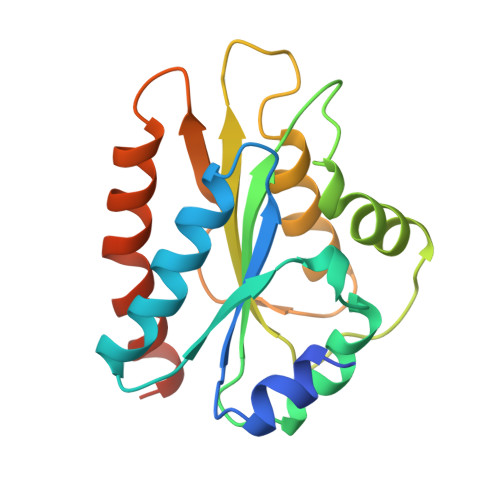
Crystal structure of the FMN-binding domain of human cytochrome P450 reductase at 1.93 A resolution.
Zhao, Q., Modi, S., Smith, G., Paine, M., McDonagh, P.D., Wolf, C.R., Tew, D., Lian, L.Y., Roberts, G.C., Driessen, H.P.(1999) Protein Sci 8: 298-306
- PubMed: 10048323
- DOI: https://doi.org/10.1110/ps.8.2.298
- Primary Citation of Related Structures:
1B1C - PubMed Abstract:
The crystal structure of the FMN-binding domain of human NADPH-cytochrome P450 reductase (P450R-FMN), a key component in the cytochrome P450 monooxygenase system, has been determined to 1.93 A resolution and shown to be very similar both to the global fold in solution (Barsukov I et al., 1997, J Biomol NMR 10:63-75) and to the corresponding domain in the 2.6 A crystal structure of intact rat P450R (Wang M et al., 1997, Proc Nat Acad Sci USA 94:8411-8416). The crystal structure of P450R-FMN reported here confirms the overall similarity of its alpha-beta-alpha architecture to that of the bacterial flavodoxins, but reveals differences in the position, number, and length of the helices relative to the central beta-sheet. The marked similarity between P450R-FMN and flavodoxins in the interactions between the FMN and the protein, indicate a striking evolutionary conservation of the FMN binding site. The P450R-FMN molecule has an unusual surface charge distribution, leading to a very strong dipole, which may be involved in docking cytochrome P450 into place for electron transfer near the FMN. Several acidic residues near the FMN are identified by mutagenesis experiments to be important for electron transfer to P4502D6 and to cytochrome c, a clear indication of the part of the molecular surface that is likely to be involved in substrate binding. Somewhat different parts are found to be involved in binding cytochrome P450 and cytochrome c.
- ICRF Unit of Structural Molecular Biology, Department of Crystallography, Birkbeck College, London, United Kingdom.
Organizational Affiliation: