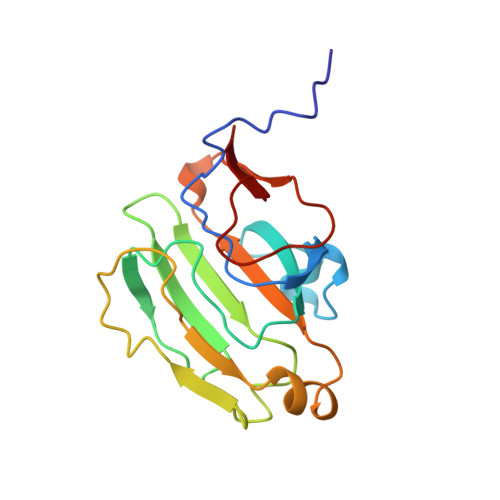
High-resolution solution structure of Bacillus subtilis IIAglc.
Chen, Y., Case, D.A., Reizer, J., Saier Jr., M.H., Wright, P.E.(1998) Proteins 31: 258-270
- PubMed: 9593197
- Primary Citation of Related Structures:
1AX3 - PubMed Abstract:
The high-resolution solution structure of the phosphocarrier protein IIAglc from Bacillus subtilis is determined using 3D and 4D heteronuclear NMR methods. B. subtilis IIAglc contains 162 amino acid residues and is one of the larger proteins for which high-resolution solution structure has been determined by NMR methods. The structures have been calculated from a total of 2,232 conformational constraints. Comparison with the X-ray crystal structure indicates that the overall fold is the same in solution and in crystalline environments, although some local structural differences are observed. These occur largely in turns and loops, and mostly correspond to regions with high-temperature factors in the crystal structure. The N-terminus of IIAglc is disordered in solution. The active site is located in a concave region of the protein surface. The histidine, which accepts the phosphoryl group (His 83), interacts with a neighboring histidine (His 68) and is surrounded by hydrophobic residues.
- Department of Molecular Biology, The Scripps Research Institute, La Jolla, California 92037, USA.
Organizational Affiliation: