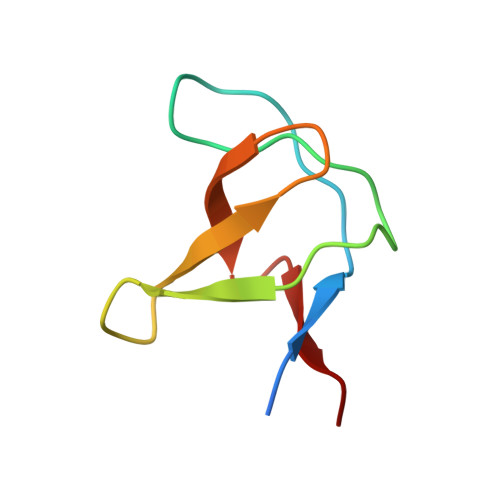
The solution structure of Abl SH3, and its relationship to SH2 in the SH(32) construct.
Gosser, Y.Q., Zheng, J., Overduin, M., Mayer, B.J., Cowburn, D.(1995) Structure 3: 1075-1086
- PubMed: 8590002
- DOI: https://doi.org/10.1016/s0969-2126(01)00243-x
- Primary Citation of Related Structures:
1AWO - PubMed Abstract:
The Src homology domains, SH3 and SH2, of Abl protein tyrosine kinase regulate enzymatic activity in vivo. Abl SH3 suppresses kinase activity, whereas Abl SH2 is required for the transforming activity of the activated form of Abl. We expect that the solution structures of Abl SH3, Abl SH2 and Abl SH(32) (a dual domain comprising SH3 and SH2 subdomains) will contribute to a structural basis for understanding the mechanism of the Abl 'regulatory apparatus'. We present the solution structure of the free Abl SH3 domain and a structural characterization of the Abl regulatory apparatus, the SH(32) dual domain. The solution structure of Abl SH3 was determined using multidimensional double resonance NMR spectroscopy. It consists of two antiparallel beta sheets packed orthogonally, an arrangement first shown in spectrin SH3. Compared with the crystal structure of the Abl SH3 complexed with a natural ligand, there is no significant difference in overall folding pattern. The structure of the Abl SH(32) dual domain was characterized by NMR spectroscopy using the 1H and 15N resonance assignment of Abl SH3 and Abl SH2. On the basis of the high degree of similarity in chemical shifts and hydrogen/deuterium exchange pattern for the individual domains of SH3 and SH2 compared with those of the SH(32) dual domain, a structural model of the Abl SH(32) regulatory apparatus is suggested. This model is in good agreement with the ligand-binding characteristics of Abl SH3, SH2 and SH(32). The binding constants for isolated SH3 and SH2 domains when binding to natural ligands, measured by intrinsic fluorescence quenching, do not differ significantly from the constants of these domains within SH(32). The solution structures of free Abl SH3 and Abl SH2, and the structural model of Abl SH(32), provide information about the overall topology of these modular domains. The structural model of Abl SH(32), a monomer, consists of the SH3 and SH2 domains connected by a flexible linker. Sites of ligand binding for the two subdomains are independent.
- Rockefeller University, New York, NY 10021, USA.
Organizational Affiliation: