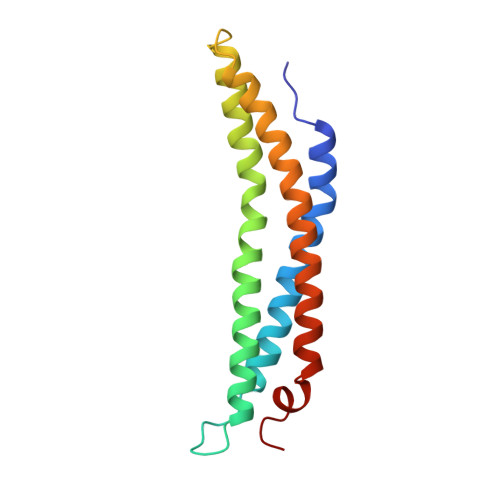
Structure of the proteasome activator REGalpha (PA28alpha).
Knowlton, J.R., Johnston, S.C., Whitby, F.G., Realini, C., Zhang, Z., Rechsteiner, M., Hill, C.P.(1997) Nature 390: 639-643
- PubMed: 9403698
- DOI: https://doi.org/10.1038/37670
- Primary Citation of Related Structures:
1AVO - PubMed Abstract:
The specificity of the 20S proteasome, which degrades many intracellular proteins, is regulated by protein complexes that bind to one or both ends of the cylindrical proteasome structure. One of these regulatory complexes, the 11S regulator (known as REG or PA28), stimulates proteasome peptidase activity and enhances the production of antigenic peptides for presentation by class I molecules of the major histocompatibility complex (MHC). The three REG subunits that have been identified, REGalpha, REGbeta and REGgamma (also known as the Ki antigen), share extensive sequence similarity, apart from a highly variable internal segment of 17-34 residues which may confer subunit-specific properties. REGalpha and REGbeta preferentially form a heteromeric complex, although purified REGalpha forms a heptamer in solution and has biochemical properties similar to the heteromeric REGalpha/REGbeta complex. We have now determined the crystal structure of human recombinant REGalpha at 2.8 A resolution. The heptameric barrel-shaped assembly contains a central channel that has an opening of 20 A diameter at one end and another of 30 A diameter at the presumed proteasome-binding surface. The binding of REG probably causes conformational changes that open a pore in the proteasome alpha-subunits through which substrates and products can pass.
- Department of Biochemistry, University of Utah, Salt Lake City 84132, USA.
Organizational Affiliation: