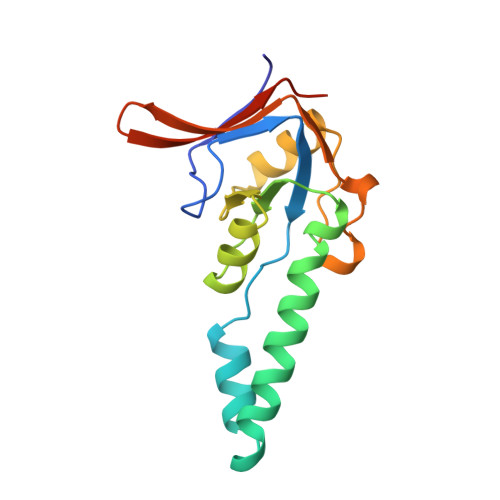
Structure of the substrate binding domain of the thermosome, an archaeal group II chaperonin.
Klumpp, M., Baumeister, W., Essen, L.O.(1997) Cell 91: 263-270
- PubMed: 9346243
- DOI: https://doi.org/10.1016/s0092-8674(00)80408-0
- Primary Citation of Related Structures:
1ASS, 1ASX - PubMed Abstract:
The crystal structure of the substrate binding domain of the thermosome, the archaeal group II chaperonin, has been determined at 2.3 A resolution. The core resembles the apical domain of GroEL but lacks the hydrophobic residues implied in binding of substrates to group I chaperonins. Rather, a large hydrophobic surface patch is found in a novel helix-turn-helix motif, which is characteristic of all group II chaperonins including the eukaryotic TRiC/CCT complex. Models of the holochaperonin, which are consistent with cryo electron microscopy data, suggest a dual role of this helical protrusion in substrate binding and controlling access to the central cavity independent of a GroES-like cochaperonin.
- Department of Molecular Structural Biology, Max-Planck-Institute for Biochemistry, Planegg-Martinsried, Germany.
Organizational Affiliation: