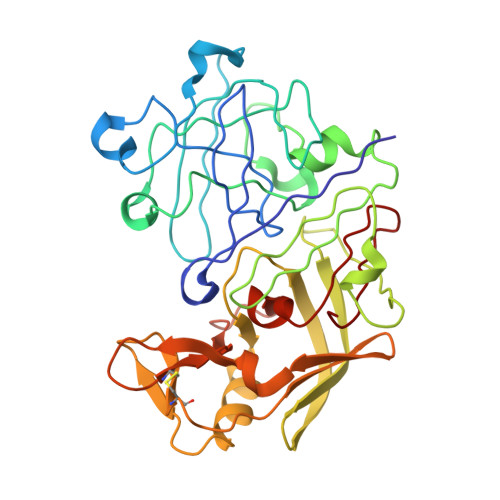
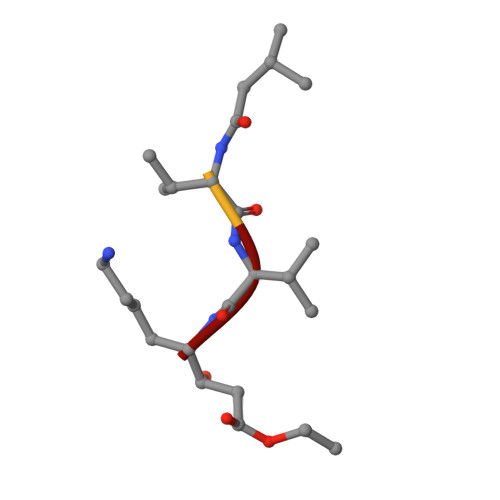
Crystallographic Analysis of a Pepstatin Analogue Binding to the Aspartyl Proteinase Penicillopepsin at 1.8 Angstroms Resolution
James, M.N.G., Sielecki, A.R., Moult, J.(1983) Peptides: Structure And Function, Proceedings Of The Of The Eighth American Peptide Symposium 1: 521