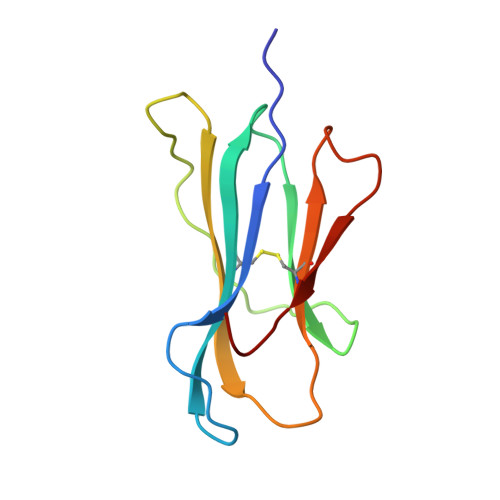
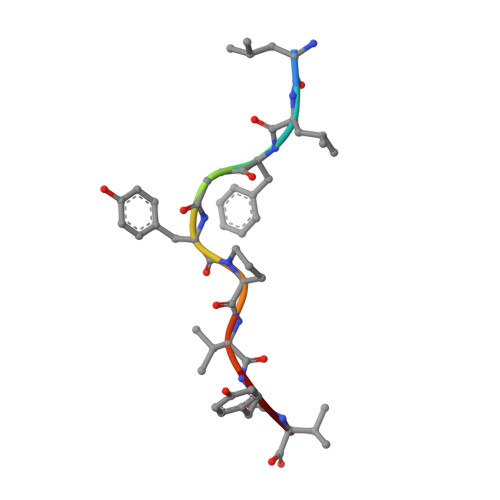
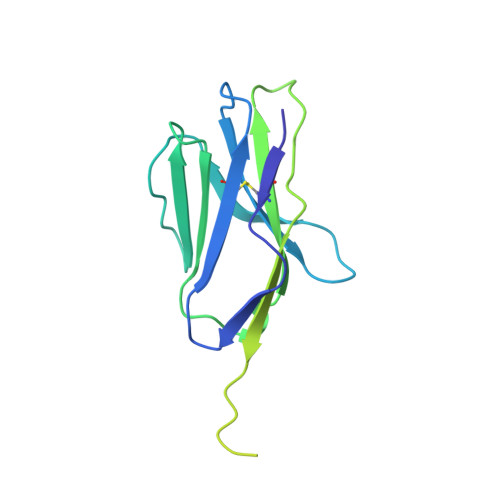
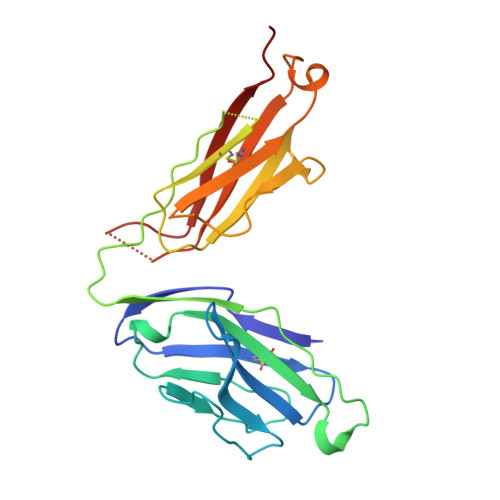
Structure of the complex between human T-cell receptor, viral peptide and HLA-A2.
Garboczi, D.N., Ghosh, P., Utz, U., Fan, Q.R., Biddison, W.E., Wiley, D.C.(1996) Nature 384: 134-141
- PubMed: 8906788
- DOI: https://doi.org/10.1038/384134a0
- Primary Citation of Related Structures:
1AO7 - PubMed Abstract:
Recognition by a T-cell antigen receptor (TCR) of peptide complexed with a major histocompatibility complex (MHC) molecule occurs through variable loops in the TCR structure which bury almost all the available peptide and a much larger area of the MHC molecule. The TCR fits diagonally across the MHC peptide-binding site in a surface feature common to all class I and class II MHC molecules, providing evidence that the nature of binding is general. A broadly applicable binding mode has implications for the mechanism of repertoire selection and the magnitude of alloreactions.
- Department of Molecular and Cellular Biology, Harvard University, Cambridge, Massachusetts 02138, USA.
Organizational Affiliation: