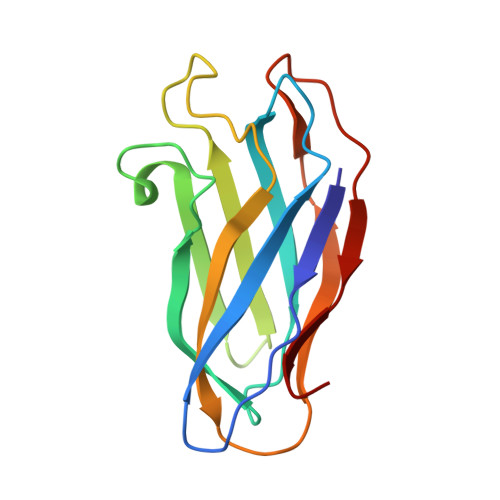
A cohesin domain from Clostridium thermocellum: the crystal structure provides new insights into cellulosome assembly.
Shimon, L.J., Bayer, E.A., Morag, E., Lamed, R., Yaron, S., Shoham, Y., Frolow, F.(1997) Structure 5: 381-390
- PubMed: 9083107
- DOI: https://doi.org/10.1016/s0969-2126(97)00195-0
- Primary Citation of Related Structures:
1ANU - PubMed Abstract:
The scaffoldin component of the cellulolytic bacterium Clostridium thermocellum is a non-hydrolytic protein which organizes the hydrolytic enzymes in a large complex, called the cellulosome. Scaffoldin comprises a series of functional domains, amongst which is a single cellulose-binding domain and nine cohesin domains which are responsible for integrating the individual enzymatic subunits into the complex. The cohesin domains are highly conserved in their primary amino acid sequences. These domains interact with a complementary domain, termed the dockerin domain, one of which is located on each enzymatic subunit. The cohesin-dockerin interaction is the crucial interaction for complex formation in the cellulosome. The determination of structural information about the cohesin domain will provide insights into cellulosome assembly and activity. We have determined the three-dimensional crystal structure of one of the cohesin domains from C. thermocellum (cohesin 2) at 2.15 A resolution. The domain forms a nine-stranded beta sandwich with a jelly-roll topology, somewhat similar to the fold displayed by its neighboring cellulose-binding domain. The compact nature of the cohesin structure and its lack of a defined binding pocket suggests that binding between the cohesin and dockerin domains is characterized by interactions between exposed surface residues. As the cohesin-dockerin interaction appears to be rather nonselective, the binding face would presumably be characterized by surface residues which exhibit both intraspecies conservation and interspecies dissimilarity. Within the same species, unconserved surface residues may reflect the position of a given cohesin domain within the scaffoldin subunit, its orientation and interactions with neighboring domains.
- Faculty of Chemistry, The Weizmann Institute of Science, Rehovot 76100, Israel. cvlinda@weizmann.weizmann.ac.il
Organizational Affiliation: