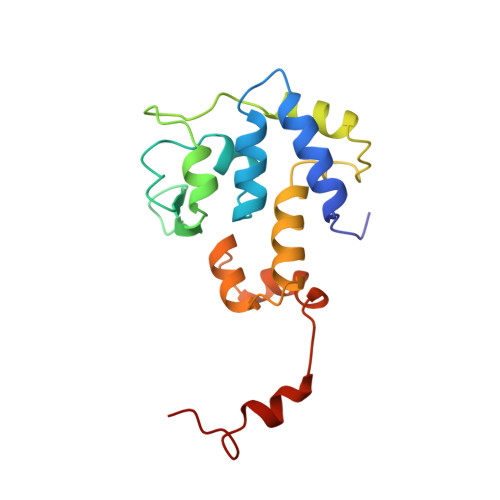
Molecular organization in site-specific recombination: the catalytic domain of bacteriophage HP1 integrase at 2.7 A resolution.
Hickman, A.B., Waninger, S., Scocca, J.J., Dyda, F.(1997) Cell 89: 227-237
- PubMed: 9108478
- DOI: https://doi.org/10.1016/s0092-8674(00)80202-0
- Primary Citation of Related Structures:
1AIH - PubMed Abstract:
HP1 integrase promotes site-specific recombination of the HP1 genome into that of Haemophilus influenzae. The isolated C-terminal domain (residues 165-337) of the protein interacts with the recombination site and contains the four catalytic residues conserved in the integrase family. This domain represents a novel fold consisting principally of well-packed alpha helices, a surface beta sheet, and an ordered 17-residue C-terminal tail. The conserved triad of basic residues and the active-site tyrosine are contributed by a single monomer and occupy fixed positions in a defined active-site cleft. Dimers are formed by mutual interactions of the tail of one monomer with an adjacent monomer; this orients active-site clefts antiparallel to each other.
- Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, Maryland 20892, USA.
Organizational Affiliation: