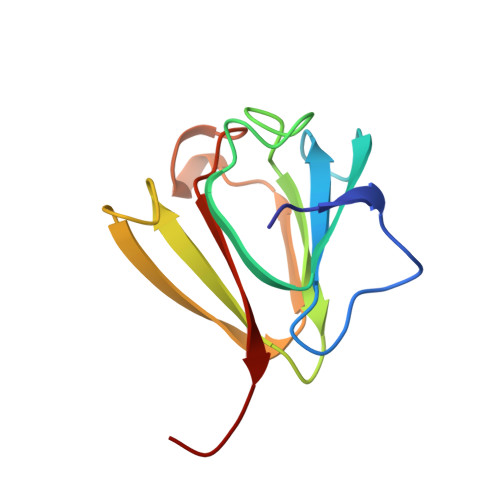
Ca2+-loaded spherulin 3a from Physarum polycephalum adopts the prototype gamma-crystallin fold in aqueous solution.
Rosinke, B., Renner, C., Mayr, E.M., Jaenicke, R., Holak, T.A.(1997) J Mol Biology 271: 645-655
- PubMed: 9281431
- DOI: https://doi.org/10.1006/jmbi.1997.1184
- Primary Citation of Related Structures:
1AG4 - PubMed Abstract:
Spherulin 3a is the most abundantly expressed cytosolic protein in spherulating plasmodia of the slime mold Physarum polycephalum. High yields of unlabeled, uniformly 15N and uniformly 13C/15N-labeled recombinant spherulin 3a from Escherichia coli could be produced by a simple protocol described here. The three-dimensional solution structure of Ca2+-loaded spherulin 3a was determined by homo- and heteronuclear NMR spectroscopy. The structure of monomeric spherulin 3a consists of two pleated beta-sheets plus a short alpha-helix arranged into the gamma-crystallin fold. The beta-sheets comprise two intertwined Greek-key motifs. An additional N-terminal beta-strand is unique to spherulin 3a. Complexation of calcium ions greatly enhances overall conformational stability of the protein. The average atomic root-mean-square deviations (r.m.s.d.) for heavy atoms in beta-strands were 0.34(+/-0.16) A for the backbone atoms and 0.73(+/-0.40) A for all atoms. The corresponding r.m.s.d. values for heavy atoms in the whole protein were 0.62(+/-0.42) A for the backbone atoms and 0.99(+/-0.65) A for all atoms. We show the structural relationship between spherulin 3a, a myxomycete dormancy protein, and crystallins from the vertebrate eye lens. Since spherulin 3a has a structure corresponding to one domain of bovine gammaB(II)-crystallin, it represents a hypothetical ancestral gamma-crystallin precursor structure.
- Max-Planck-Institut für Biochemie, Martinsried, D-82152, Germany.
Organizational Affiliation: