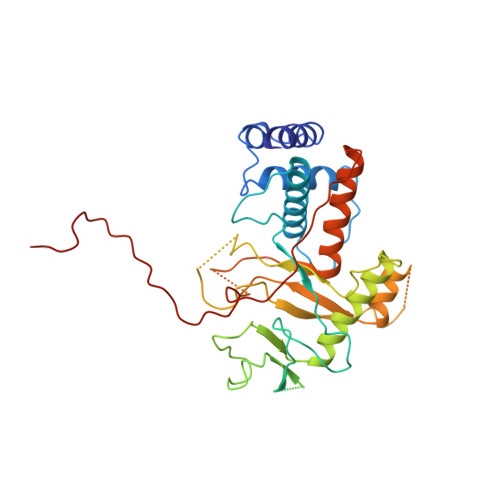
Alternative arrangements of the protein chain are possible for the adenovirus single-stranded DNA binding protein.
Kanellopoulos, P.N., Tsernoglou, D., van der Vliet, P.C., Tucker, P.A.(1996) J Mol Biology 257: 1-8
- PubMed: 8632448
- DOI: https://doi.org/10.1006/jmbi.1996.0141
- Primary Citation of Related Structures:
1ADU, 1ADV - PubMed Abstract:
A second crystal form of the C-terminal domain of the adenovirus single-stranded DNA binding protein crystallizes in space group P2(1)2(1)2(1) with a=61.0 angstrom, b=91.2 angstrom and c=149.4 angstrom. The crystals contain two molecules per asymmetric unit and diffract to a maximum resolution of 3.0 angstrom. The crystal is composed of infinite chains of molecules along the crystallographic 2(1) axis parallel to c. The principal intermolecular interaction is a hooking of the C-terminal 17 residues of one molecule onto the next molecule in the protein chain. Adjacent molecules in the chain are rotated approximately 90 degrees with respect to their neighbours. The difference in relative orientation of adjacent molecules between the two crystal forms of the protein implies a degree of flexibility in the protein chain that would facilitate DNA binding.
- Structural Biology Programme, European Molecular Biology Laboratory, Heidelberg, Germany.
Organizational Affiliation: