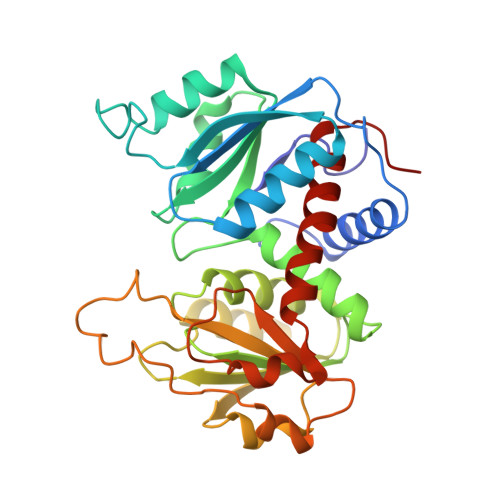
Arginine 54 in the active site of Escherichia coli aspartate transcarbamoylase is critical for catalysis: a site-specific mutagenesis, NMR, and X-ray crystallographic study.
Stebbins, J.W., Robertson, D.E., Roberts, M.F., Stevens, R.C., Lipscomb, W.N., Kantrowitz, E.R.(1992) Protein Sci 1: 1435-1446
- PubMed: 1303763
- DOI: https://doi.org/10.1002/pro.5560011105
- Primary Citation of Related Structures:
1ACM - PubMed Abstract:
The replacement of Arg-54 by Ala in the active site of Escherichia coli aspartate transcarbamoylase causes a 17,000-fold loss of activity but does not significantly influence the binding of substrates or substrate analogs (Stebbins, J.W., Xu, W., & Kantrowitz, E.R., 1989, Biochemistry 28, 2592-2600). In the X-ray structure of the wild-type enzyme, Arg-54 interacts with both the anhydride oxygen and a phosphate oxygen of carbamoyl phosphate (CP) (Gouaux, J.E. & Lipscomb, W.N., 1988, Proc. Natl. Acad. Sci. USA 85, 4205-4208). The Arg-54-->Ala enzyme was crystallized in the presence of the transition state analog N-phosphonacetyl-L-aspartate (PALA), data were collected to a resolution limit of 2.8 A, and the structure was solved by molecular replacement. The analysis of the refined structure (R factor = 0.18) indicates that the substitution did not cause any significant alterations to the active site, except that the side chain of the arginine was replaced by two water molecules. 31P-NMR studies indicate that the binding of CP to the wild-type catalytic subunit produces an upfield chemical shift that cannot reflect a significant change in the ionization state of the CP but rather indicates that there are perturbations in the electronic environment around the phosphate moiety when CP binds to the enzyme. The pH dependence of this upfield shift for bound CP indicates that the catalytic subunit undergoes a conformational change with a pKa approximately 7.7 upon CP binding. Furthermore, the linewidth of the 31P signal of CP bound to the Arg-54-->Ala enzyme is significantly narrower than that of CP bound to the wild-type catalytic subunit at any pH, although the change in chemical shift for the CP bound to the mutant enzyme is unaltered. 31P-NMR studies of PALA complexed to the wild-type catalytic subunit indicate that the phosphonate group of the bound PALA exists as the dianion at pH 7.0 and 8.8, whereas in the Arg-54-->Ala catalytic subunit the phosphonate group of the bound PALA exists as the monoanion at pH 7.0 and 8.8. Thus, the side chain of Arg-54 is essential for the proper ionization of the phosphonate group of PALA and by analogy the phosphate group in the transition state. These data support the previously proposed proton transfer mechanism, in which a fully ionized phosphate group in the transition state accepts a proton during catalysis.
- Department of Chemistry, Merkert Chemistry Center, Boston College, Chestnut Hill, Massachusetts 02167.
Organizational Affiliation: