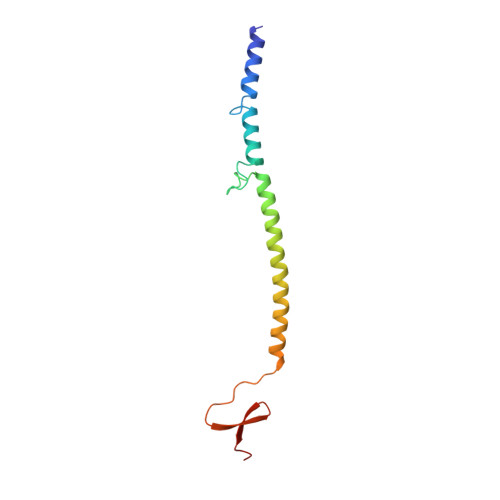
Structure of bacteriophage T4 fibritin: a segmented coiled coil and the role of the C-terminal domain.
Tao, Y., Strelkov, S.V., Mesyanzhinov, V.V., Rossmann, M.G.(1997) Structure 5: 789-798
- PubMed: 9261070
- DOI: https://doi.org/10.1016/s0969-2126(97)00233-5
- Primary Citation of Related Structures:
1AA0 - PubMed Abstract:
Oligomeric coiled-coil motifs are found in numerous protein structures; among them is fibritin, a structural protein of bacteriophage T4, which belongs to a class of chaperones that catalyze a specific phage-assembly process. Fibritin promotes the assembly of the long tail fibers and their subsequent attachment to the tail baseplate; it is also a sensing device that controls the retraction of the long tail fibers in adverse environments and, thus, prevents infection. The structure of fibritin had been predicted from sequence and biochemical analyses to be mainly a triple-helical coiled coil. The determination of its structure at atomic resolution was expected to give insights into the assembly process and biological function of fibritin, and the properties of modified coiled-coil structures in general. The three-dimensional structure of fibritin E, a deletion mutant of wild-type fibritin, was determined to 2.2 A resolution by X-ray crystallography. Three identical subunits of 119 amino acid residues form a trimeric parallel coiled-coil domain and a small globular C-terminal domain about a crystallographic threefold axis. The coiled-coil domain is divided into three segments that are separated by insertion loops. The C-terminal domain, which consists of 30 residues from each subunit, contains a beta-propeller-like structure with a hydrophobic interior. The residues within the C-terminal domain make extensive hydrophobic and some polar intersubunit interactions. This is consistent with the C-terminal domain being important for the correct assembly of fibritin, as shown earlier by mutational studies. Tight interactions between the C-terminal residues of adjacent subunits counteract the latent instability that is suggested by the structural properties of the coiled-coil segments. Trimerization is likely to begin with the formation of the C-terminal domain which subsequently initiates the assembly of the coiled coil. The interplay between the stabilizing effect of the C-terminal domain and the labile coiled-coil domain may be essential for the fibritin function and for the correct functioning of many other alpha-fibrous proteins.
- Department of Biological Sciences, Purdue University, West Lafayette, Indiana 47907-1392, USA.
Organizational Affiliation: