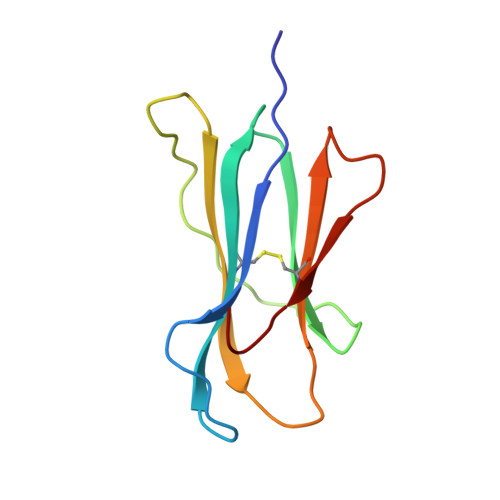
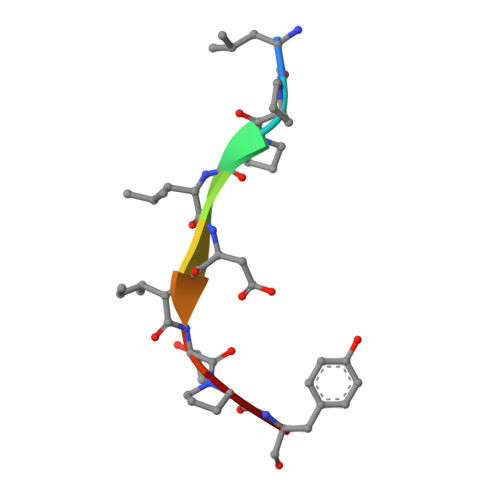
Decamer-like conformation of a nona-peptide bound to HLA-B*3501 due to non-standard positioning of the C terminus.
Menssen, R., Orth, P., Ziegler, A., Saenger, W.(1999) J Mol Biology 285: 645-653
- PubMed: 9878435
- DOI: https://doi.org/10.1006/jmbi.1998.2363
- Primary Citation of Related Structures:
1A9B, 1A9E - PubMed Abstract:
The N and C termini of peptides presented by major histocompatibility complex (MHC) class I molecules are held within the peptide binding groove by a network of hydrogen bonds to conserved MHC residues. However, the published structure of the human allele HLA-B*3501 complexed with the nef octa-peptide VPLRPMTY, revealed non-standard positioning for both peptide termini. To investigate whether these deviations are indeed related to the length of the nef-peptide, we have determined the structure of HLA-B*3501 presenting a nona-peptide to 2.5 A resolution. A comparison of HLA-B*3501/peptide complexes with structures of other HLA molecules exhibits allele-specific properties of HLA-B*3501, as well as peptide-induced structural changes. Independent of the length of the bound peptide, HLA-B*3501 positions the peptide C terminus significantly closer to the alpha1-helix and nearer to the A pocket than observed for other HLA class I/peptide complexes. This reorientation is accompanied by a shift within the N-terminal part of the alpha2-helix towards the middle of the binding groove. Due to the short distance between the N and C termini, the nona-peptide is compressed and forced to zig-zag vertically within the binding groove. Its conformation rather resembles that of a deca-peptide than of other nona-peptides bound to class I molecules. Superposition of both HLA-B*3501/peptide complexes additionally reveals a significant, peptide-dependent deviation between the N-terminal parts of the alpha1-helices which might be due to different positioning of the peptide N termini. Taken together, these data illustrate the strong interdependence between the HLA class I molecule and the bound peptide.
- Institut für Immungenetik Universitätsklinikum Charité, Humboldt-Universität zu Berlin, Spandauer Damm 130, Berlin, D-14050, Germany.
Organizational Affiliation: