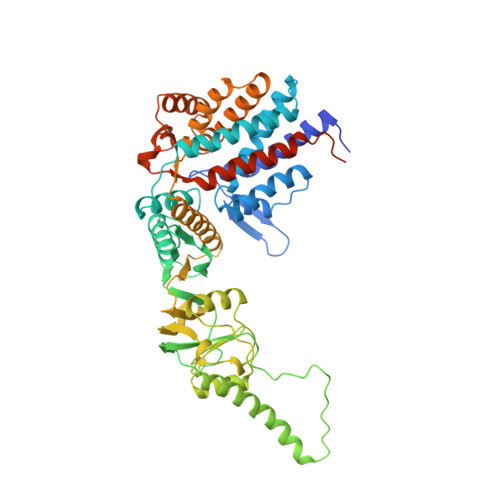
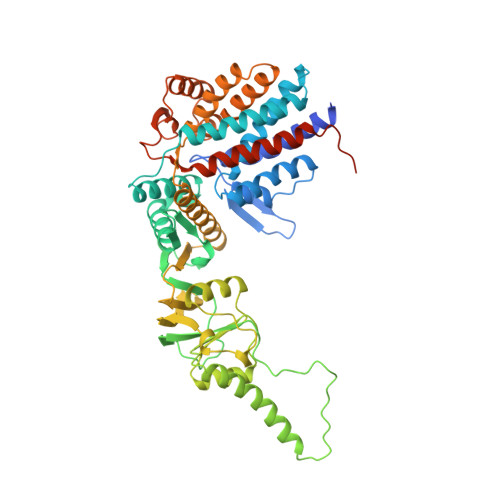
Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT.
Ditzel, L., Lowe, J., Stock, D., Stetter, K.O., Huber, H., Huber, R., Steinbacher, S.(1998) Cell 93: 125-138
- PubMed: 9546398
- DOI: https://doi.org/10.1016/s0092-8674(00)81152-6
- Primary Citation of Related Structures:
1A6D, 1A6E - PubMed Abstract:
We have determined to 2.6 A resolution the crystal structure of the thermosome, the archaeal group II chaperonin from T. acidophilum. The hexadecameric homolog of the eukaryotic chaperonin CCT/TRiC shows an (alphabeta)4(alphabeta)4 subunit assembly. Domain folds are homologous to GroEL but form a novel type of inter-ring contact. The domain arrangement resembles the GroEL-GroES cis-ring. Parts of the apical domains form a lid creating a closed conformation. The lid substitutes for a GroES-like cochaperonin that is absent in the CCT/TRiC system. The central cavity has a polar surface implicated in protein folding. Binding of the transition state analog Mg-ADP-AIF3 suggests that the closed conformation corresponds to the ATP form.
- Max-Planck-Institut für Biochemie, Martinsried, Germany.
Organizational Affiliation: