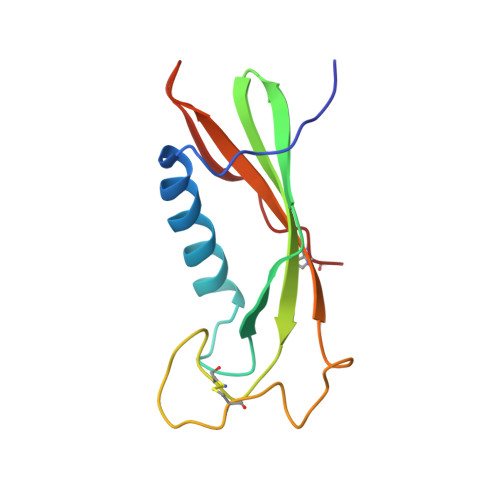
The structures of native phosphorylated chicken cystatin and of a recombinant unphosphorylated variant in solution.
Dieckmann, T., Mitschang, L., Hofmann, M., Kos, J., Turk, V., Auerswald, E.A., Jaenicke, R., Oschkinat, H.(1993) J Mol Biology 234: 1048-1059
- PubMed: 8263912
- DOI: https://doi.org/10.1006/jmbi.1993.1658
- Primary Citation of Related Structures:
1A67, 1A90 - PubMed Abstract:
The solution structures of the phosphorylated form of native chicken cystatin and the recombinant variant AEF-S1M-M29I-M89L were determined by 2D, 3D and 4D-NMR. The structures turn out to be very similar, despite the substitutions and the phosphorylation of the wild-type. Their dominant feature is a five-stranded beta-sheet, which is wrapped around a five-turn alpha-helix, as shown by X-ray crystallographic studies of wild-type chicken cystatin. However, the NMR analysis shows that the second helix observed in the crystal is not present in solution. The phosphorylation occurs at S80, which is located in a flexible region. For this reason, very few effects on the structure are observed. Comparison of structures of the unphosphorylated variant and the wild-type shows small effects on H84 which is located in the supposed recognition site of the serine kinase. This recognition site appears to be well structured as a large loop-containing bulge of the beta-sheet. The N termini of both mutants, which contribute to a large extent to the binding to the proteinase, are very flexible. A loop structure involving the residues L7 to A10 as found in related inhibitors, such as in the kininogen domains 2 and 3, is not sufficiently populated to be observed.
- Max-Planck-Institut für Biochemie, Martinsried bei München, Germany.
Organizational Affiliation: