The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions.
Das, A.K., Cohen, P.W., Barford, D.(1998) EMBO J 17: 1192-1199
- PubMed: 9482716
- DOI: https://doi.org/10.1093/emboj/17.5.1192
- Primary Citation of Related Structures:
1A17 - PubMed Abstract:
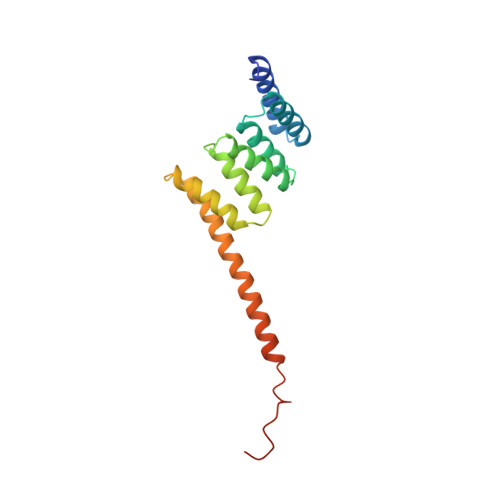
The tetratricopeptide repeat (TPR) is a degenerate 34 amino acid sequence identified in a wide variety of proteins, present in tandem arrays of 3-16 motifs, which form scaffolds to mediate protein-protein interactions and often the assembly of multiprotein complexes. TPR-containing proteins include the anaphase promoting complex (APC) subunits cdc16, cdc23 and cdc27, the NADPH oxidase subunit p67 phox, hsp90-binding immunophilins, transcription factors, the PKR protein kinase inhibitor, and peroxisomal and mitochondrial import proteins. Here, we report the crystal structure of the TPR domain of a protein phosphatase, PP5. Each of the three TPR motifs of this domain consist of a pair of antiparallel alpha-helices of equivalent length. Adjacent TPR motifs are packed together in a parallel arrangement such that a tandem TPR motif structure is composed of a regular series of antiparallel alpha-helices. The uniform angular and spatial arrangement of neighbouring alpha-helices defines a helical structure and creates an amphipathic groove. Multiple-TPR motif proteins would fold into a right-handed super-helical structure with a continuous helical groove suitable for the recognition of target proteins, hence defining a novel mechanism for protein recognition. The spatial arrangement of alpha-helices in the PP5-TPR domain is similar to those within 14-3-3 proteins.
- Laboratory of Molecular Biophysics, University of Oxford, Rex Richards Building, South Parks Road, Oxford, OX1 3QU, UK.
Organizational Affiliation: