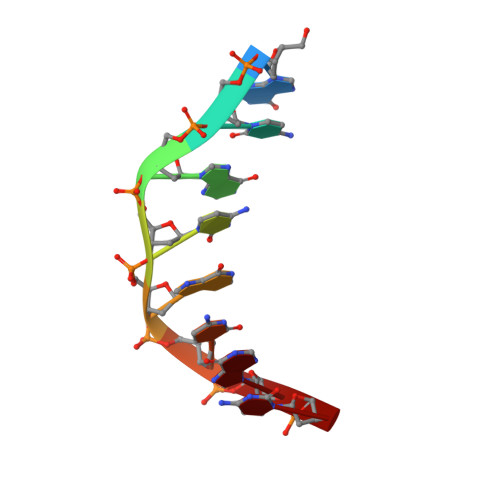
Binding of two distamycin A molecules in the minor groove of an alternating B-DNA duplex.
Chen, X., Ramakrishnan, B., Rao, S.T., Sundaralingam, M.(1994) Nat Struct Biol 1: 169-175
- PubMed: 7656035
- DOI: https://doi.org/10.1038/nsb0394-169
- Primary Citation of Related Structures:
159D - PubMed Abstract:
Here we report the 1.8A X-ray structure of a 2:1 drug-DNA complex between distamycin A and an alternating B-DNA octamer duplex d(ICICICIC)2. The two distamycin A molecules are bound side by side with dyad symmetry in an antiparallel orientation in the expanded minor groove. The amides of each drug molecule are hydrogen bonded to the minor groove base atoms of only one DNA strand. The complex not only shows binding of two drug molecules, but the DNA duplex also exhibits striking low-high alternations in the helical twist angles, the sugar puckering and the phosphate conformations, providing the basis for a new model for an alternating B-DNA with a dinucleotide repeat.
- Department of Chemistry, Ohio State University, Columbus 43210, USA.
Organizational Affiliation: