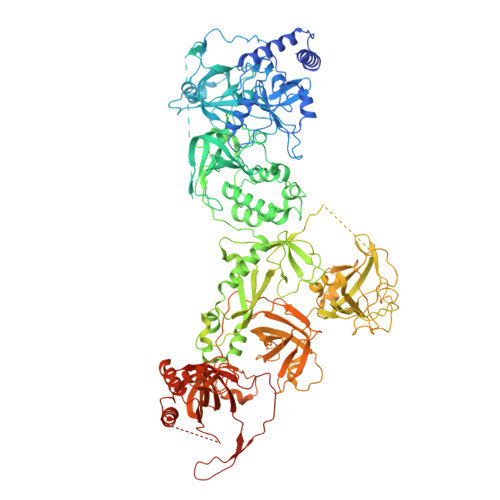
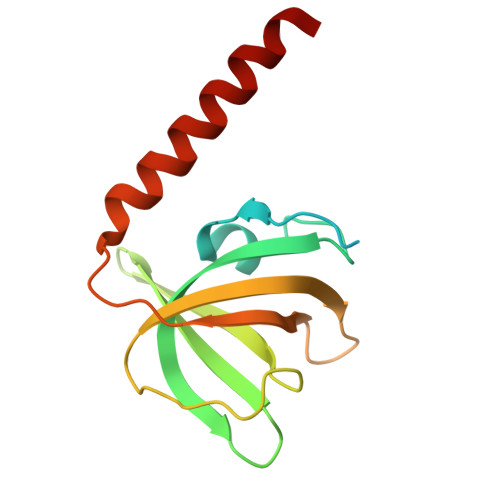
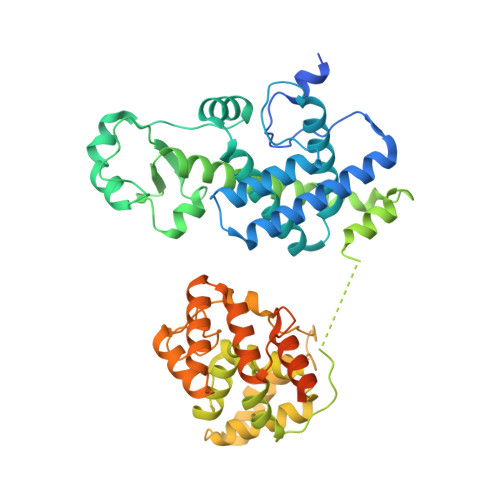
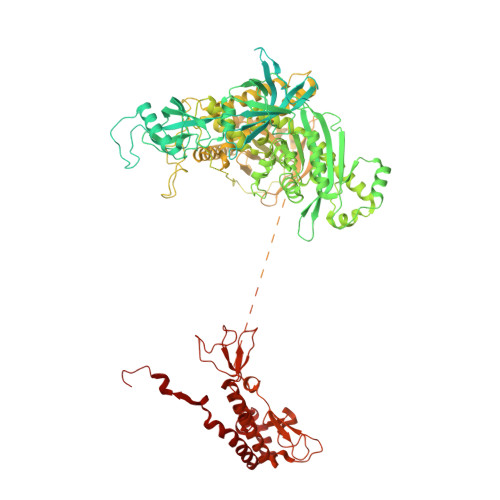
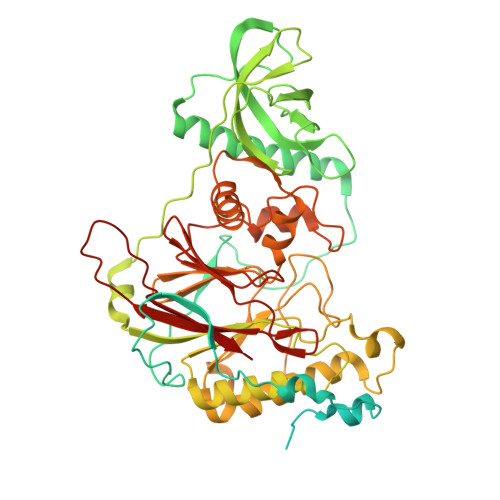
Structures of the human CST-Pol alpha-primase complex bound to telomere templates.
He, Q., Lin, X., Chavez, B.L., Agrawal, S., Lusk, B.L., Lim, C.J.(2022) Nature 608: 826-832
- PubMed: 35830881
- DOI: https://doi.org/10.1038/s41586-022-05040-1
- Primary Citation of Related Structures:
8D0B, 8D0K - PubMed Abstract:
The mammalian DNA polymerase-α-primase (Polα-primase) complex is essential for DNA metabolism, providing the de novo RNA-DNA primer for several DNA replication pathways 1-4 such as lagging-strand synthesis and telomere C-strand fill-in. The physical mechanism underlying how Polα-primase, alone or in partnership with accessory proteins, performs its complicated multistep primer synthesis function is unknown. Here we show that CST, a single-stranded DNA-binding accessory protein complex for Polα-primase, physically organizes the enzyme for efficient primer synthesis. Cryogenic electron microscopy structures of the CST-Polα-primase preinitiation complex (PIC) bound to various types of telomere overhang reveal that template-bound CST partitions the DNA and RNA catalytic centres of Polα-primase into two separate domains and effectively arranges them in RNA-DNA synthesis order. The architecture of the PIC provides a single solution for the multiple structural requirements for the synthesis of RNA-DNA primers by Polα-primase. Several insights into the template-binding specificity of CST, template requirement for assembly of the CST-Polα-primase PIC and activation are also revealed in this study.
Organizational Affiliation:
Department of Biochemistry, University of Wisconsin-Madison, Madison, WI, USA.