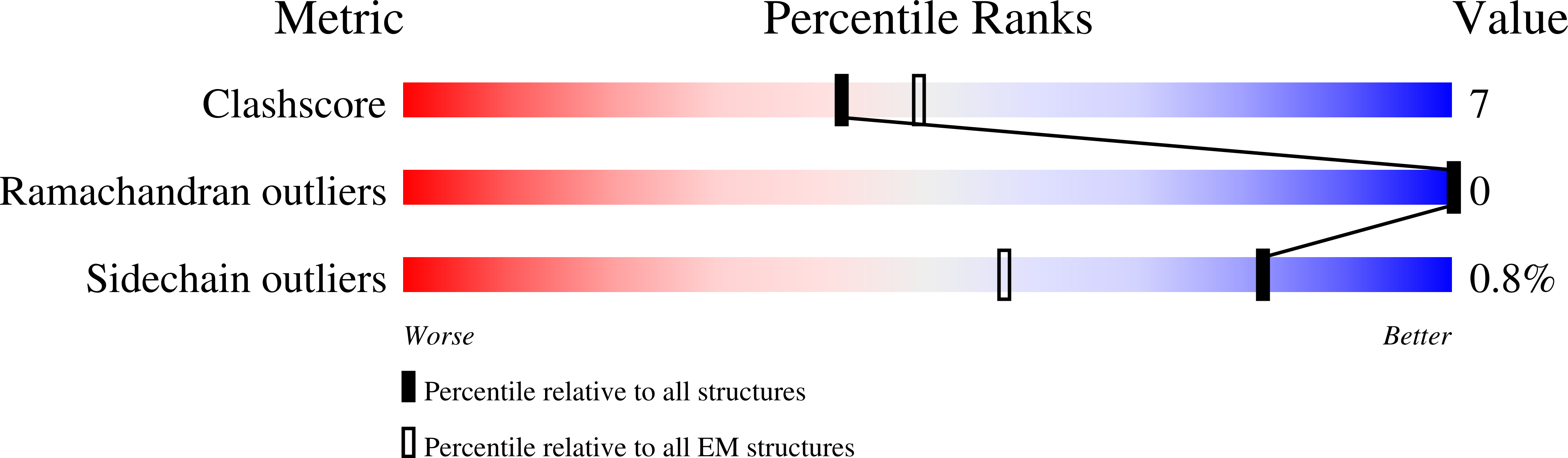Structural Insights into Common and Host-Specific Receptor-Binding Mechanisms in Algal Picorna-like Viruses.
Wang, H., Munke, A., Li, S., Tomaru, Y., Okamoto, K.(2022) Viruses 14
- PubMed: 36366467
- DOI: https://doi.org/10.3390/v14112369
- Primary Citation of Related Structures:
8B38, 8B3J - PubMed Abstract:
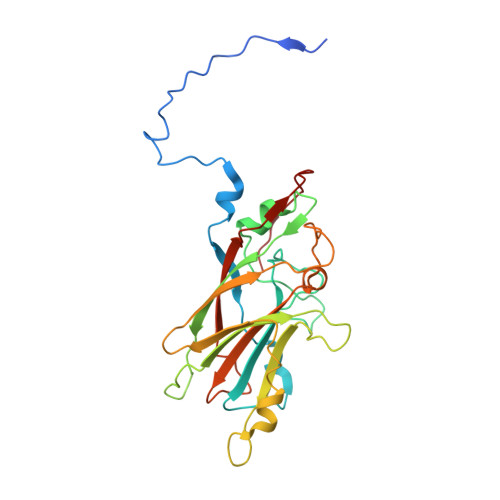
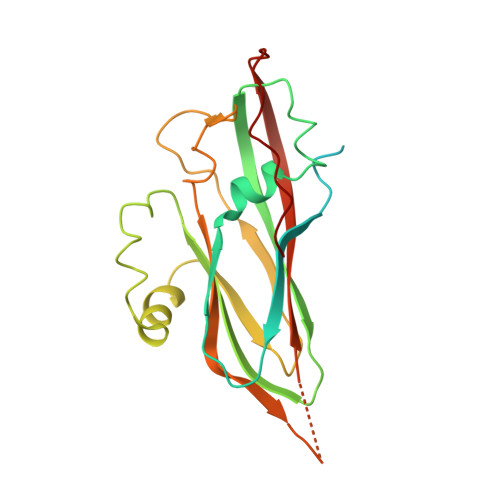
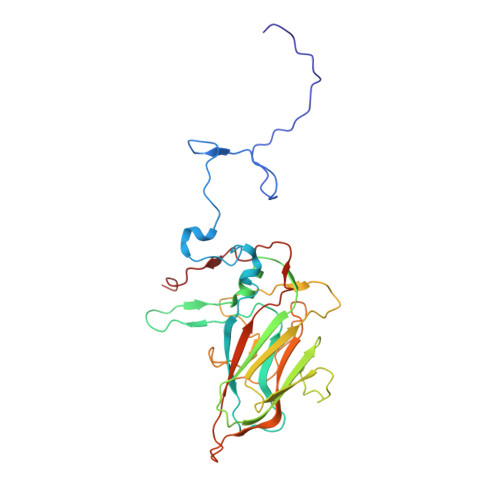
Marnaviridae viruses are abundant algal viruses that regulate the dynamics of algal blooms in aquatic environments. They employ a narrow host range because they merely lyse their algal host species. This host-specific lysis is thought to correspond to the unique receptor-binding mechanism of the Marnaviridae viruses. Here, we present the atomic structures of the full and empty capsids of Chaetoceros socialis forma radians RNA virus 1 built-in 3.0 Å and 3.1 Å cryo-electron microscopy maps. The empty capsid structure and the structural variability provide insights into its assembly and uncoating intermediates. In conjunction with the previously reported atomic model of the Chaetoceros tenuissimus RNA virus type II capsid, we have identified the common and diverse structural features of the VP1 surface between the Marnaviridae viruses. We have also tested the potential usage of AlphaFold2 for structural prediction of the VP1s and a subsequent structural phylogeny for classifying Marnaviridae viruses by their hosts. These findings will be crucial for inferring the host-specific receptor-binding mechanism in Marnaviridae viruses.
Organizational Affiliation:
The Laboratory of Molecular Biophysics, Department of Cell and Molecular Biology, Uppsala University, 75124 Uppsala, Sweden.