A Novel Lipase from Lasiodiplodia theobromae Efficiently Hydrolyses C8-C10 Methyl Esters for the Preparation of Medium-Chain Triglycerides' Precursors.
Ng, A.M.J., Yang, R., Zhang, H., Xue, B., Yew, W.S., Nguyen, G.K.T.(2021) Int J Mol Sci 22
- PubMed: 34638680
- DOI: https://doi.org/10.3390/ijms221910339
- Primary Citation of Related Structures:
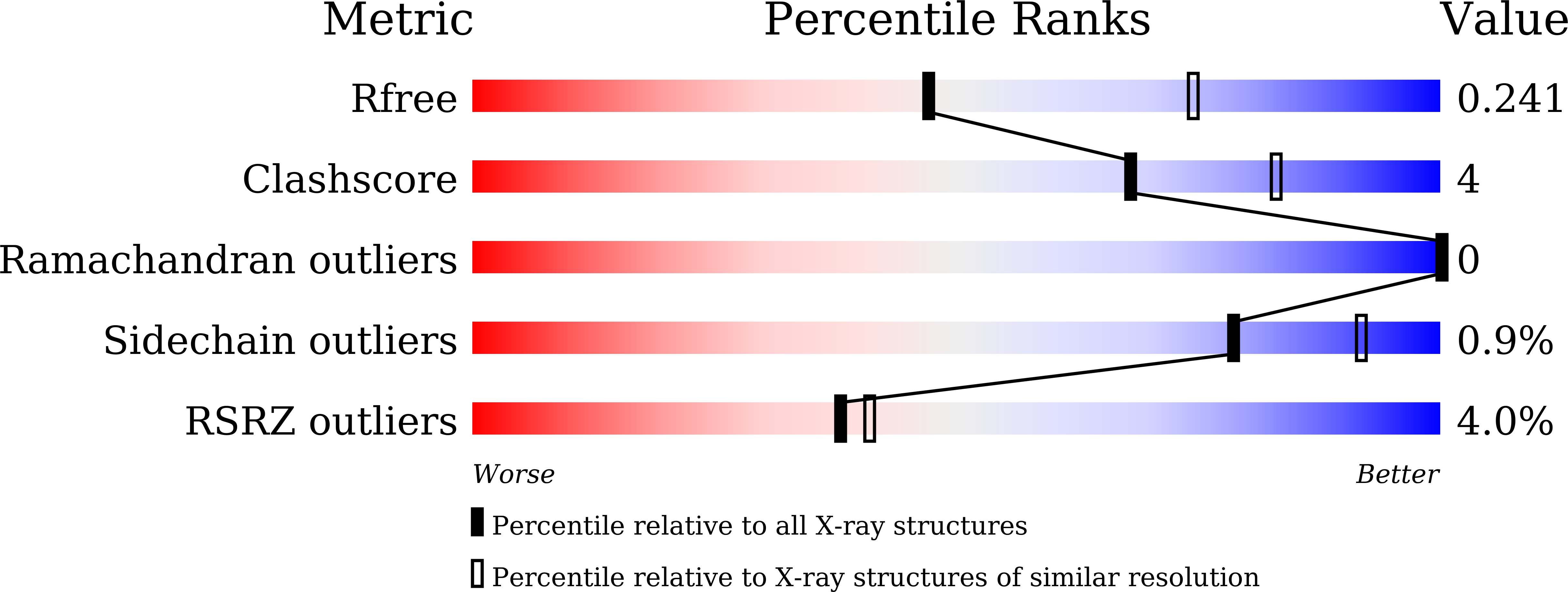
7V6D - PubMed Abstract:
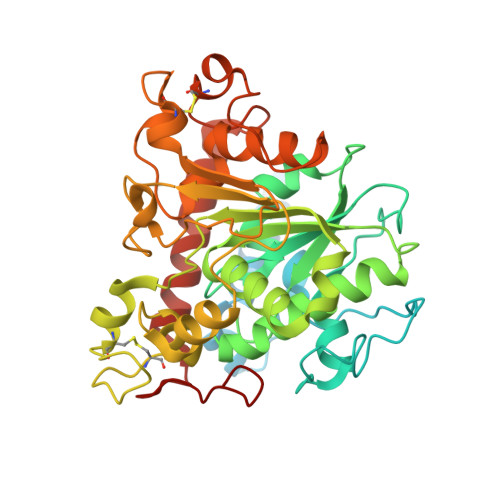
Medium-chain triglycerides (MCTs) are an emerging choice to treat neurodegenerative disorders such as Alzheimer's disease. They are triesters of glycerol and three medium-chain fatty acids, such as capric (C8) and caprylic (C10) acids. The availability of C8-C10 methyl esters (C8-C10 ME) from vegetable oil processes has presented an opportunity to use methyl esters as raw materials for the synthesis of MCTs. However, there are few reports on enzymes that can efficiently hydrolyse C8-C10 ME to industrial specifications. Here, we report the discovery and identification of a novel lipase from Lasiodiplodia theobromae fungus (LTL1), which hydrolyses C8-C10 ME efficiently. LTL1 can perform hydrolysis over pH ranges from 3.0 to 9.0 and maintain thermotolerance up to 70 °C. It has high selectivity for monoesters over triesters and displays higher activity over commercially available lipases for C8-C10 ME to achieve 96.17% hydrolysis within 31 h. Structural analysis by protein X-ray crystallography revealed LTL1's well-conserved lipase core domain, together with a partially resolved N-terminal subdomain and an inserted loop, which may suggest its hydrolytic preference for monoesters. In conclusion, our results suggest that LTL1 provides a tractable route towards to production of C8-C10 fatty acids from methyl esters for the synthesis of MCTs.
Organizational Affiliation:
WIL@NUS Corporate Laboratory, Wilmar International Limited, Centre for Translational Medicine, 14 Medical Drive, Singapore 117599, Singapore.