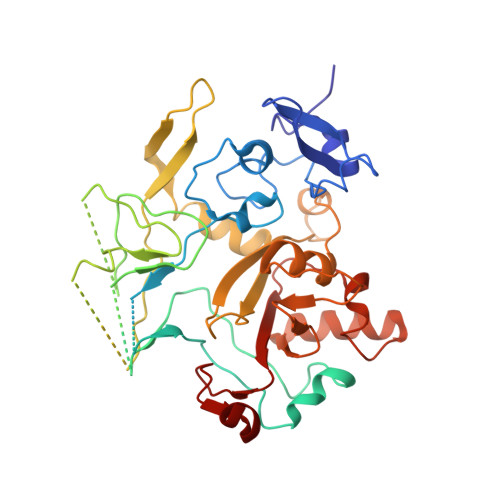
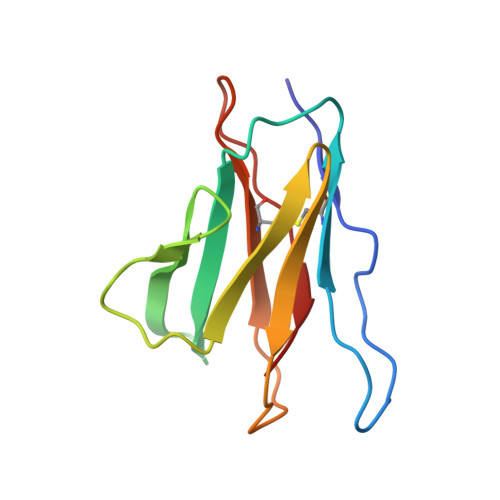
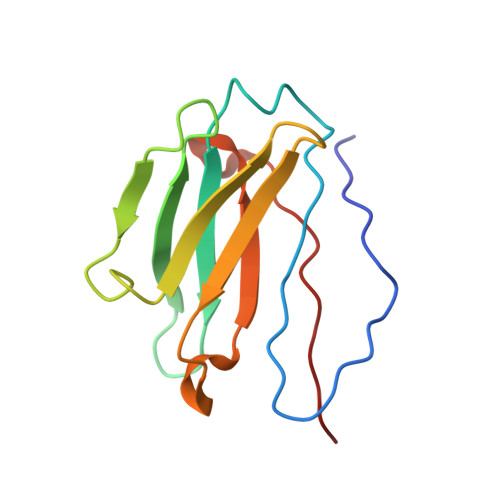
Neutralization of the anthrax toxin by antibody-mediated stapling of its membrane-penetrating loop.
Hoelzgen, F., Zalk, R., Alcalay, R., Cohen-Schwartz, S., Garau, G., Shahar, A., Mazor, O., Frank, G.A.(2021) Acta Crystallogr D Struct Biol 77: 1197-1205
- PubMed: 34473089
- DOI: https://doi.org/10.1107/S2059798321007816
- Primary Citation of Related Structures:
7O85 - PubMed Abstract:
Anthrax infection is associated with severe illness and high mortality. Protective antigen (PA) is the central component of the anthrax toxin, which is one of two major virulence factors of Bacillus anthracis, the causative agent of anthrax disease. Upon endocytosis, PA opens a pore in the membranes of endosomes, through which the cytotoxic enzymes of the toxin are extruded. The PA pore is formed by a cooperative conformational change in which the membrane-penetrating loops of PA associate, forming a hydrophobic rim that pierces the membrane. Due to its crucial role in anthrax progression, PA is an important target for monoclonal antibody-based therapy. cAb29 is a highly effective neutralizing antibody against PA. Here, the cryo-EM structure of PA in complex with the Fab portion of cAb29 was determined. It was found that cAb29 neutralizes the toxin by clamping the membrane-penetrating loop of PA to the static surface-exposed loop of the D3 domain of the same subunit, thereby preventing pore formation. These results provide the structural basis for the antibody-based neutralization of PA and bring into focus the membrane-penetrating loop of PA as a target for the development of better anti-anthrax vaccines.
Organizational Affiliation:
Department of Life Sciences, Ben-Gurion University of the Negev, Beer Sheva 8410501, Israel.