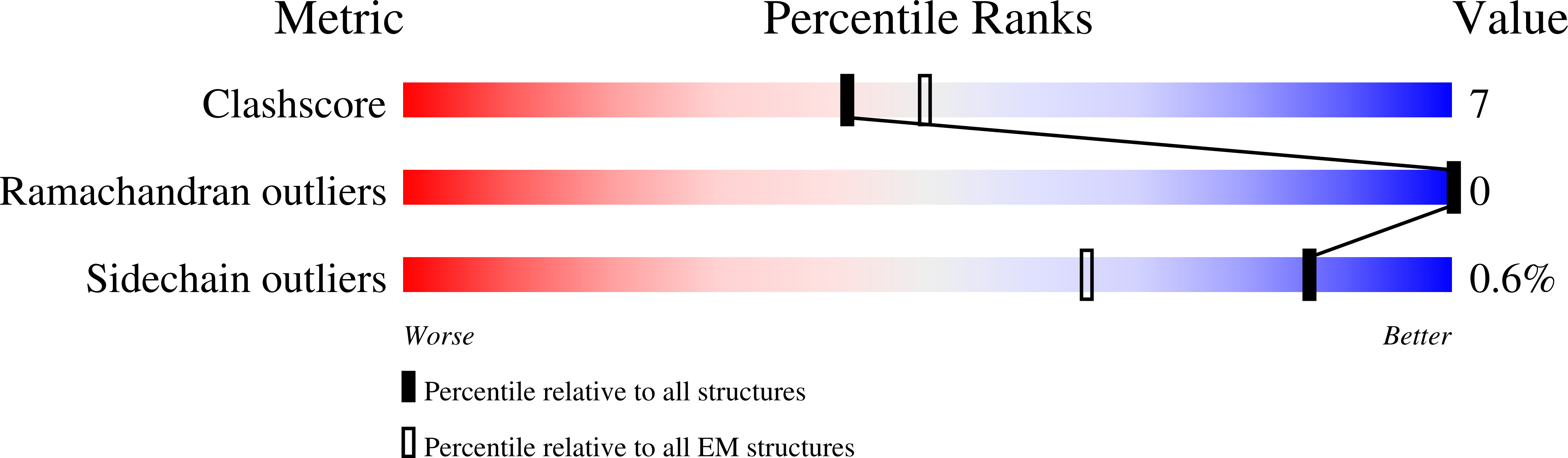Evolving cryo-EM structural approaches for GPCR drug discovery.
Zhang, X., Johnson, R.M., Drulyte, I., Yu, L., Kotecha, A., Danev, R., Wootten, D., Sexton, P.M., Belousoff, M.J.(2021) Structure 29: 963
- PubMed: 33957078
- DOI: https://doi.org/10.1016/j.str.2021.04.008
- Primary Citation of Related Structures:
7LCI, 7LCJ, 7LCK - PubMed Abstract:
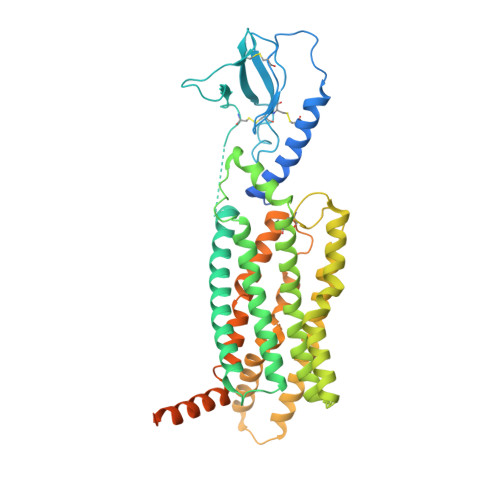
G protein-coupled receptors (GPCRs) are the largest class of cell surface drug targets. Advances in stabilization of GPCR:transducer complexes, together with improvements in cryoelectron microscopy (cryo-EM) have recently been applied to structure-assisted drug design for GPCR agonists. Nonetheless, limitations in the commercial application of these approaches, including the use of nanobody 35 (Nb35) to aid complex stabilization and the high cost of 300 kV imaging, have restricted broad application of cryo-EM in drug discovery. Here, using the PF 06882961-bound GLP-1R as exemplar, we validated the formation of stable complexes with a modified Gs protein in the absence of Nb35. In parallel, we compare 200 versus 300 kV image acquisition using a Falcon 4 or K3 direct electron detector. Moreover, the 200 kV Glacios-Falcon 4 yielded a 3.2 Å map with clear density for bound drug and multiple structurally ordered waters. Our work paves the way for broader commercial application of cryo-EM for GPCR drug discovery.
Organizational Affiliation:
Drug Discovery Biology Theme, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville 3052, VIC, Australia.