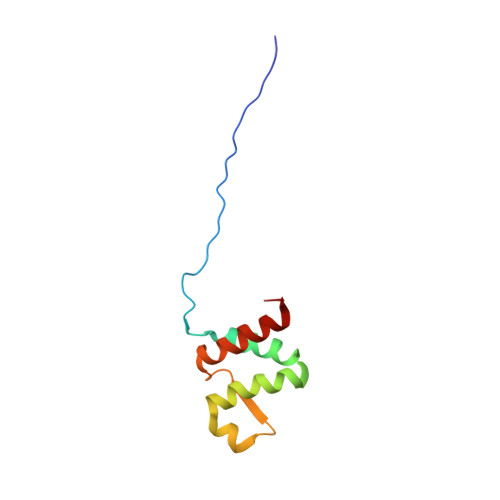
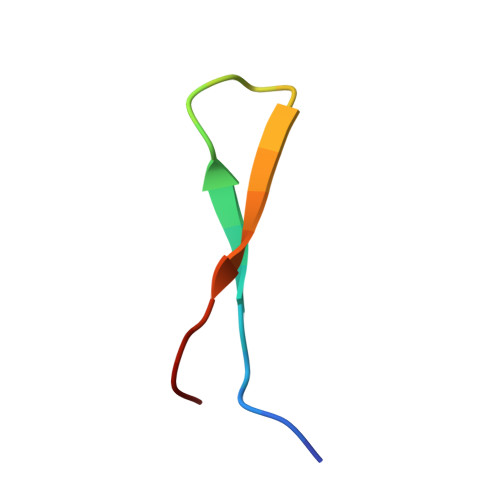
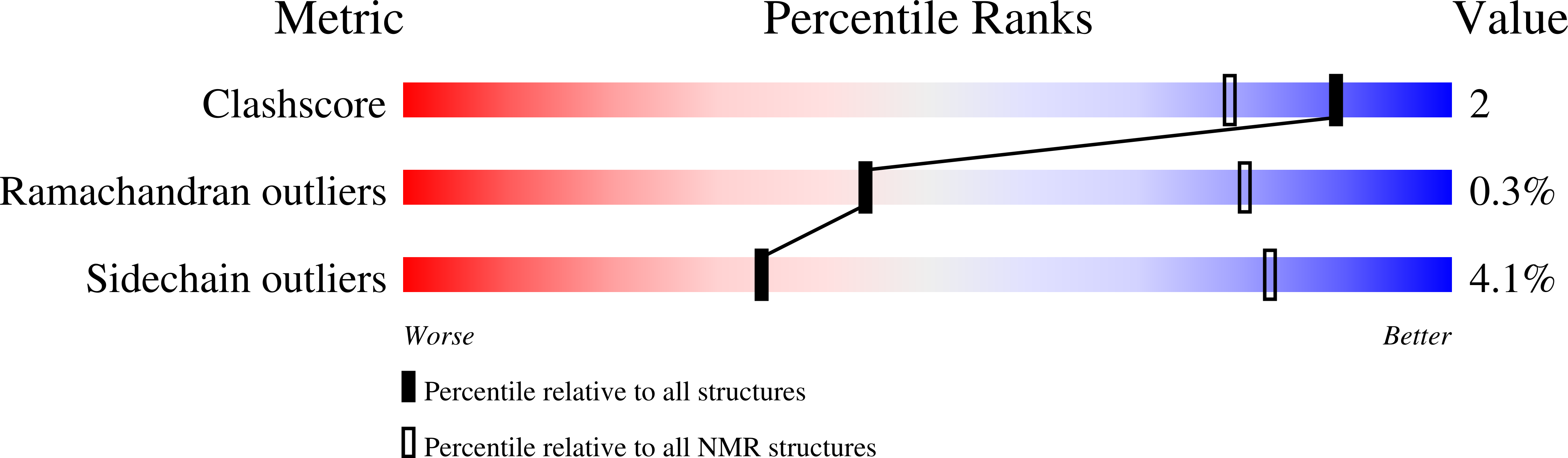A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Aiyer, S., Swapna, G.V.T., Ma, L.C., Liu, G., Hao, J., Chalmers, G., Jacobs, B.C., Montelione, G.T., Roth, M.J.(2021) Structure 29: 886
- PubMed: 33592170
- DOI: https://doi.org/10.1016/j.str.2021.01.010
- Primary Citation of Related Structures:
7JMY, 7JQ8, 7JYN, 7JYZ - PubMed Abstract:
The extraterminal (ET) domain of BRD3 is conserved among BET proteins (BRD2, BRD3, BRD4), interacting with multiple host and viral protein-protein networks. Solution NMR structures of complexes formed between the BRD3 ET domain and either the 79-residue murine leukemia virus integrase (IN) C-terminal domain (IN 329-408 ) or its 22-residue IN tail peptide (IN 386-407 ) alone reveal similar intermolecular three-stranded β-sheet formations. 15 N relaxation studies reveal a 10-residue linker region (IN 379-388 ) tethering the SH3 domain (IN 329-378 ) to the ET-binding motif (IN 389-405 ):ET complex. This linker has restricted flexibility, affecting its potential range of orientations in the IN:nucleosome complex. The complex of the ET-binding peptide of the host NSD3 protein (NSD3 148-184 ) and the BRD3 ET domain includes a similar three-stranded β-sheet interaction, but the orientation of the β hairpin is flipped compared with the two IN:ET complexes. These studies expand our understanding of molecular recognition polymorphism in complexes of ET-binding motifs with viral and host proteins.
Organizational Affiliation:
Department of Pharmacology, Robert Wood Johnson Medical School, Rutgers University, Piscataway, NJ 08854, USA; Department of Biochemistry and Molecular Biology, Robert Wood Johnson Medical School, Rutgers University, Piscataway, NJ 08854, USA.