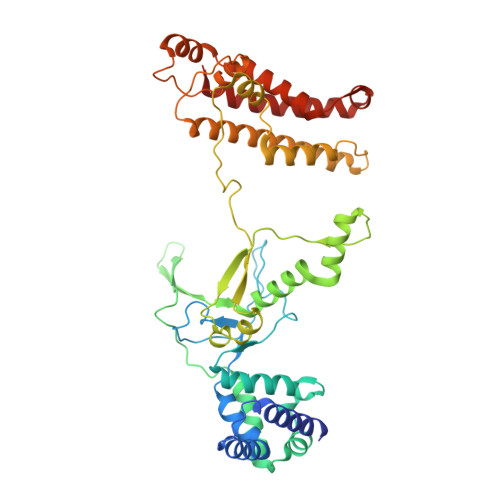
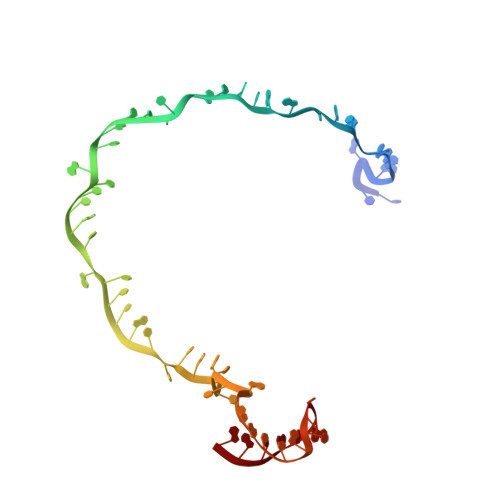
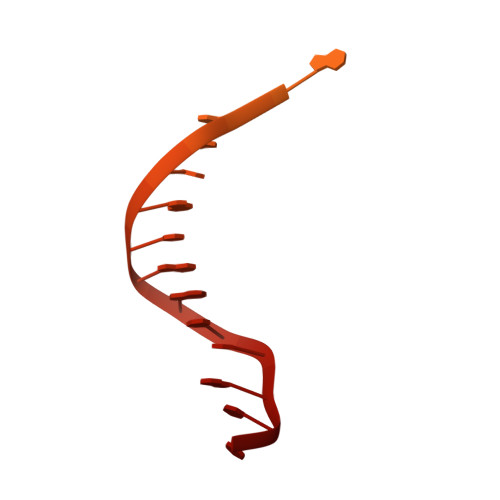
Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Yang, L., Zhang, L., Yin, P., Ding, H., Xiao, Y., Zeng, J., Wang, W., Zhou, H., Wang, Q., Zhang, Y., Chen, Z., Yang, M., Feng, Y.(2022) Nat Commun 13: 1931-1931
- PubMed: 35411005
- DOI: https://doi.org/10.1038/s41467-022-29581-1
- Primary Citation of Related Structures:
7DTR, 7ELM, 7ELN, 7WE6 - PubMed Abstract:
CRISPR-Cas systems are prokaryotic adaptive immune systems and phages use anti-CRISPR proteins (Acrs) to counteract these systems. Here, we report the structures of AcrIF24 and its complex with the crRNA-guided surveillance (Csy) complex. The HTH motif of AcrIF24 can bind the Acr promoter region and repress its transcription, suggesting its role as an Aca gene in self-regulation. AcrIF24 forms a homodimer and further induces dimerization of the Csy complex. Apart from blocking the hybridization of target DNA to the crRNA, AcrIF24 also induces the binding of non-sequence-specific dsDNA to the Csy complex, similar to AcrIF9, although this binding seems to play a minor role in AcrIF24 inhibitory capacity. Further structural and biochemical studies of the Csy-AcrIF24-dsDNA complexes and of AcrIF24 mutants reveal that the HTH motif of AcrIF24 and the PAM recognition loop of the Csy complex are structural elements essential for this non-specific dsDNA binding. Moreover, AcrIF24 and AcrIF9 display distinct characteristics in inducing non-specific DNA binding. Together, our findings highlight a multifunctional Acr and suggest potential wide distribution of Acr-induced non-specific DNA binding.
Organizational Affiliation:
Beijing Advanced Innovation Center for Soft Matter Science and Engineering, Beijing Key Laboratory of Bioprocess, State Key Laboratory of Chemical Resource Engineering, College of Life Science and Technology, Beijing University of Chemical Technology, Beijing, 100029, China.