Recognition of lipoproteins by scavenger receptor class A members.
Cheng, C., Zheng, E., Yu, B., Zhang, Z., Wang, Y., Liu, Y., He, Y.(2021) J Biol Chem 297: 100948-100948
- PubMed: 34252459
- DOI: https://doi.org/10.1016/j.jbc.2021.100948
- Primary Citation of Related Structures:
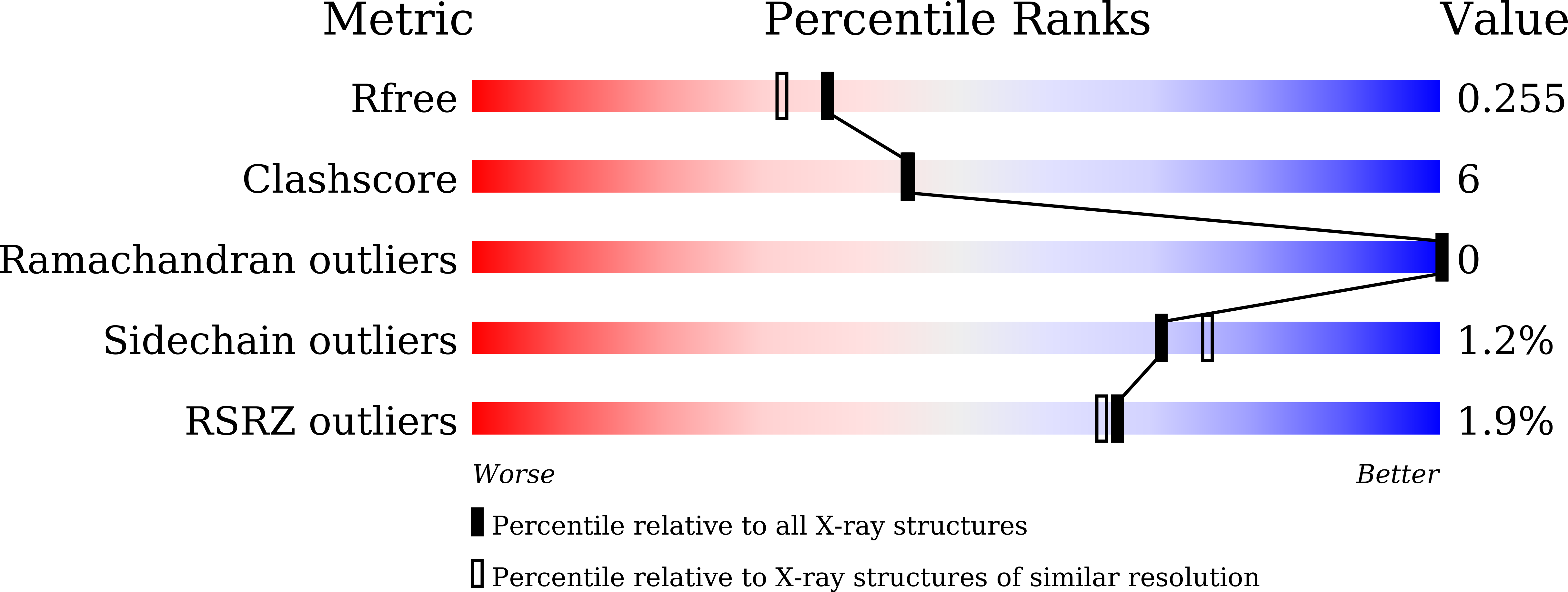
7DPX - PubMed Abstract:
Scavenger receptor class A (SR-A) proteins are type II transmembrane glycoproteins that form homotrimers on the cell surface. This family has five known members (SCARA1 to 5, or SR-A1 to A5) that recognize a variety of ligands and are involved in multiple biological pathways. Previous reports have shown that some SR-A family members can bind modified low-density lipoproteins (LDLs); however, the mechanisms of the interactions between the SR-A members and these lipoproteins are not fully understood. Here, we systematically characterize the recognition of SR-A receptors with lipoproteins and report that SCARA1 (SR-A1, CD204), MARCO (SCARA2), and SCARA5 recognize acetylated or oxidized LDL and very-low-density lipoprotein in a Ca 2+ -dependent manner through their C-terminal scavenger receptor cysteine-rich (SRCR) domains. These interactions occur specifically between the SRCR domains and the modified apolipoprotein B component of the lipoproteins, suggesting that they might share a similar mechanism for lipoprotein recognition. Meanwhile, SCARA4, a SR-A member with a carbohydrate recognition domain instead of the SRCR domain at the C terminus, shows low affinity for modified LDL and very-low-density lipoprotein but binds in a Ca 2+ -independent manner. SCARA3, which does not have a globular domain at the C terminus, was found to have no detectable binding with these lipoproteins. Taken together, these results provide mechanistic insights into the interactions between SR-A family members and lipoproteins that may help us understand the roles of SR-A receptors in lipid transport and related diseases such as atherosclerosis.
Organizational Affiliation:
State Key Laboratory of Oncogenes and Related Genes, Shanghai Cancer Institute, Renji Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China; Department of Biliary-Pancreatic Surgery, Renji Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China; Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, Shanghai, China.