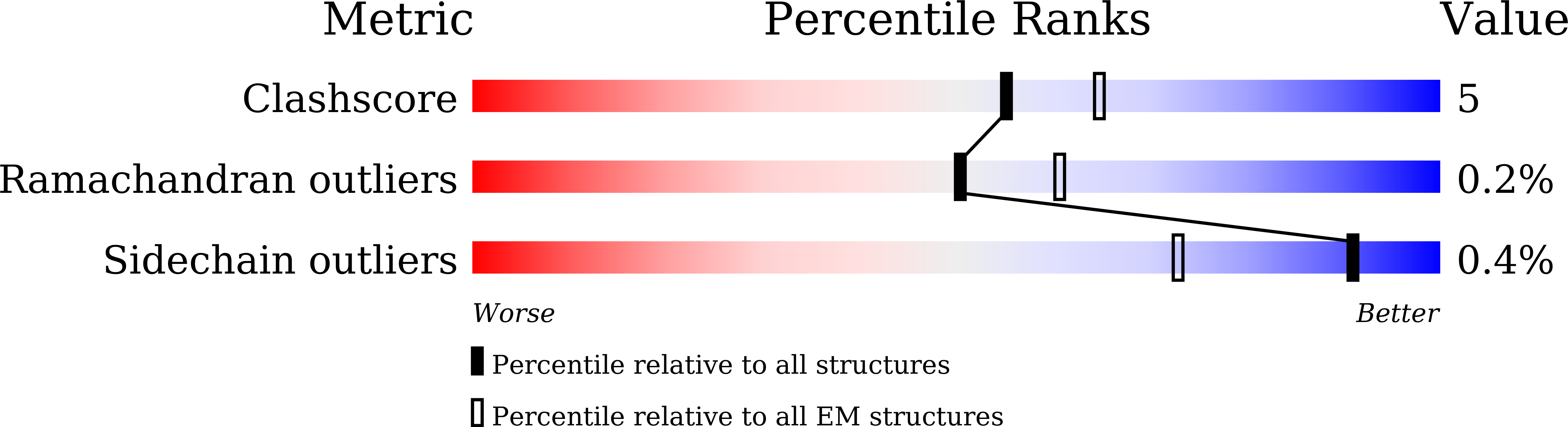
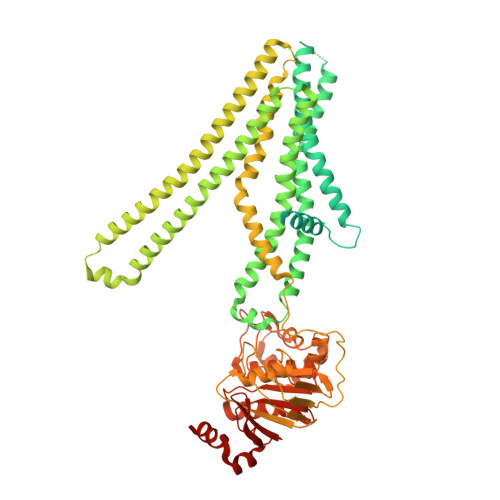
Cryo-electron microscopy structure of human ABCB6 transporter.
Wang, C., Cao, C., Wang, N., Wang, X., Wang, X., Zhang, X.C.(2020) Protein Sci 29: 2363-2374
- PubMed: 33007128
- DOI: https://doi.org/10.1002/pro.3960
- Primary Citation of Related Structures:
7D7N, 7D7R - PubMed Abstract:
Human ATP-binding cassette transporter 6 of subfamily B (ABCB6) is an ABC transporter involved in the translocation toxic metals and anti-cancer drugs. Using cryo-electron microscopy, we determined the molecular structure of full-length ABCB6 in an apo state. The structure of ABCB6 unravels the architecture of a full-length ABCB transporter that harbors two N-terminal transmembrane domains which is indispensable for its ATPase activity in our in vitro assay. A slit-like substrate binding pocket of ABCB6 may accommodate the planar shape of porphyrins, and the existence of a secondary cavity near the mitochondrial intermembrane space side would further facilitate substrate release. Furthermore, the ATPase activity of ABCB6 stimulated with a variety of porphyrin substrates showed different profiles in the presence of glutathione (GSH), suggesting the action of a distinct substrate translocation mechanism depending on the use of GSH as a cofactor.
Organizational Affiliation:
National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China.