Molecular basis for PICS-mediated piRNA biogenesis and cell division.
Wang, X., Zeng, C., Liao, S., Zhu, Z., Zhang, J., Tu, X., Yao, X., Feng, X., Guang, S., Xu, C.(2021) Nat Commun 12: 5595-5595
- PubMed: 34552083
- DOI: https://doi.org/10.1038/s41467-021-25896-7
- Primary Citation of Related Structures:
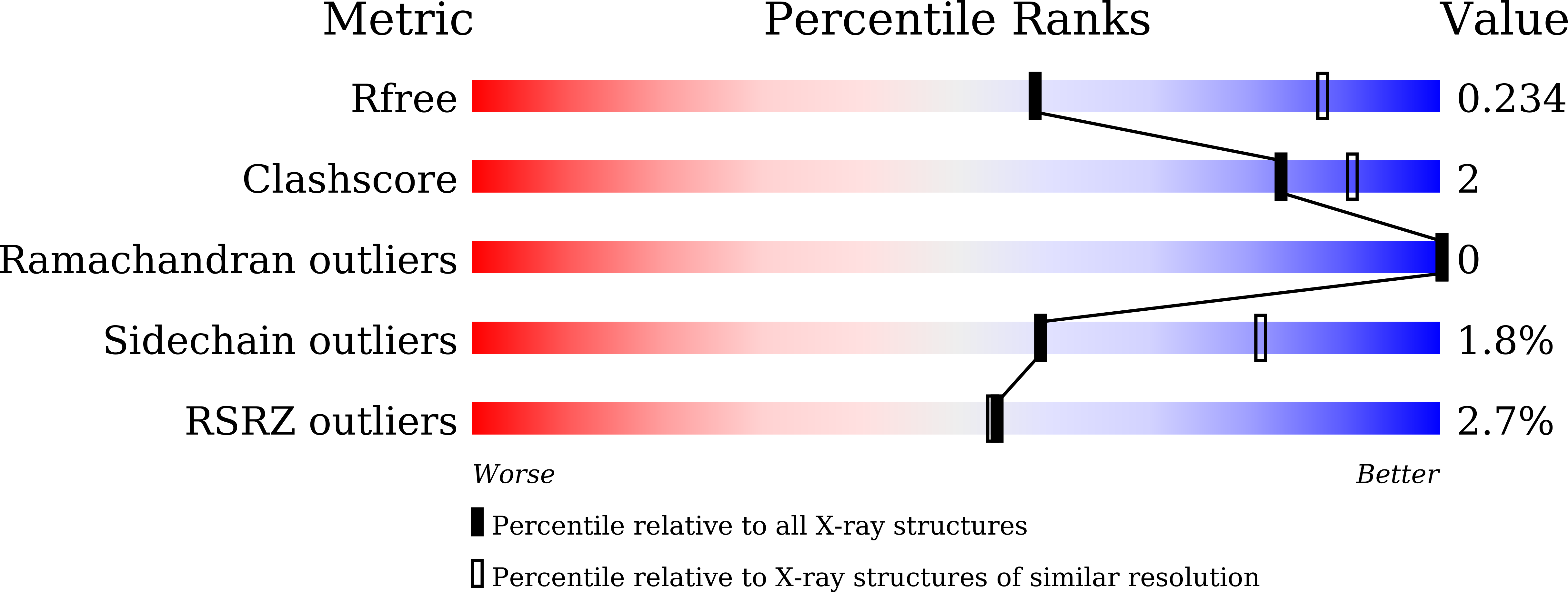
7D1L, 7D2Y, 7EJO, 7EJS - PubMed Abstract:
By incorporating two mutually exclusive factors, PID-1 and TOST-1, C. elegans PICS complex plays important roles in piRNA biogenesis, chromosome segregation and cell division. We firstly map the interaction network between PICS subunits, then uncover the mechanisms underlying the interactions between PICS subunits by solving several complex structures, including those of TOFU-6/PICS-1, ERH-2/PICS-1, and ERH-2/TOST-1. Our biochemical experiment also demonstrates that PICS exists as an octamer consisting of two copies of each subunit. Combining structural analyses with mutagenesis experiments, we identify interfacial residues of PICS subunits that are critical for maintaining intact PICS complex in vitro. Furthermore, using genetics, cell biology and imaging experiments, we find that those mutants impairing the in vitro interaction network within PICS, also lead to dysfunction of PICS in vivo, including mislocalization of PICS, and reduced levels of piRNAs or aberrant chromosome segregation and cell division. Therefore, our work provides structural insights into understanding the PICS-mediated piRNA biogenesis and cell division.
Organizational Affiliation:
Ministry of Education Key Laboratory for Membraneless Organelles & Cellular Dynamics, Hefei National Laboratory for Physical Sciences at the Microscale, School of Life Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, PR China.