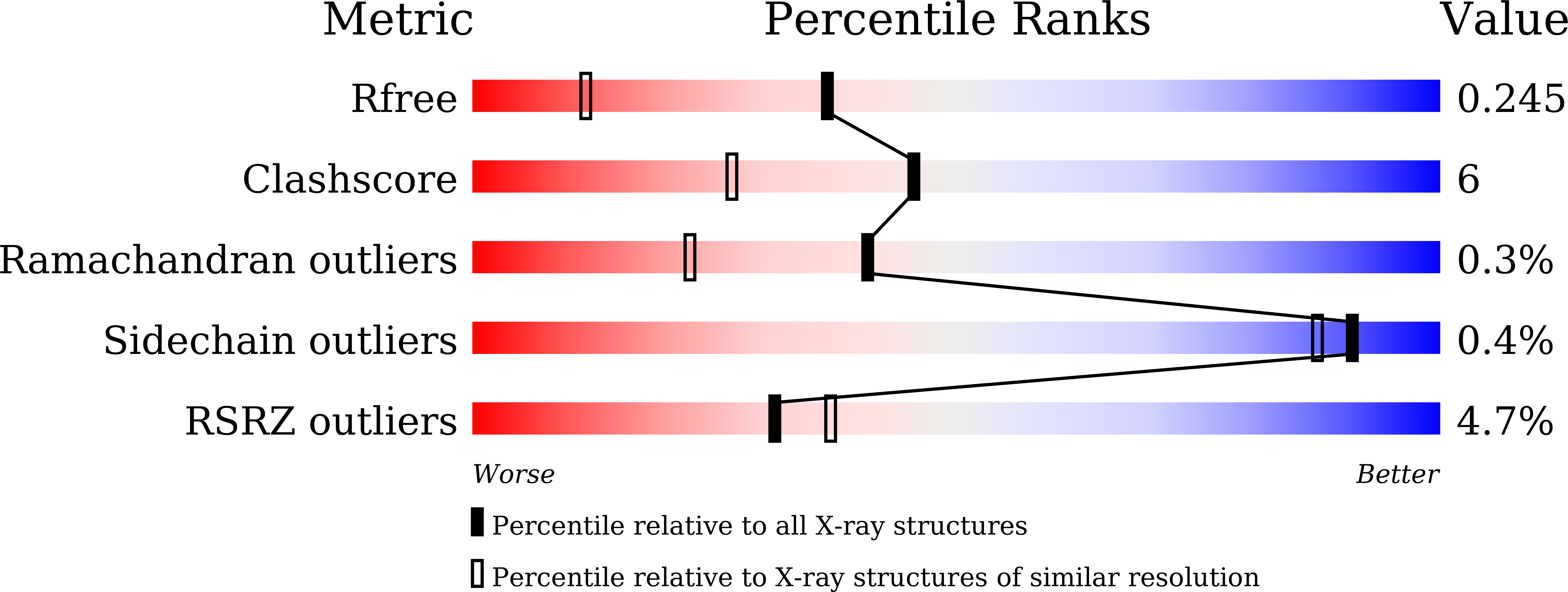
Structural insights into the multiple binding modes of Dimethyl Fumarate (DMF) and its analogs to the Kelch domain of Keap1.
Unni, S., Deshmukh, P., Krishnappa, G., Kommu, P., Padmanabhan, B.(2021) FEBS J 288: 1599-1613
- PubMed: 32672401
- DOI: https://doi.org/10.1111/febs.15485
- Primary Citation of Related Structures:
6LRZ, 7C5E, 7C60 - PubMed Abstract:
The activation of the nuclear factor erythroid 2-related factor 2 (Nrf2) transcription function has been implicated in the protection of neurodegenerative diseases. The cytoplasmic protein, Kelch-like ECH-associated protein 1 (Keap1), negatively regulates Nrf2. The Keap1-Nrf2 pathway is a potential therapeutic target for tackling free-radical damage. Dimethyl fumarate (DMF) is currently an approved drug for the treatment of relapsing multiple sclerosis. Recent studies showed that DMF modifies the reactive cysteines in the BTB domain of Keap1 and thus activates Nrf2 transcription function. Intriguingly, our crystal structure studies revealed that DMF also binds to the β-propeller domain (Keap1-DC) of Keap1. The crystal structure of the complex, refined to 1.54 Å resolution, revealed unexpected features: DMF binds (a) to the Nrf2-binding site (bottom region of Keap1-DC, site 1) with moderate interaction, and (b) to the top region of Keap1-DC, near to the blade II (site 2). The specificity of the binding 'site 2' was found to be unique to blade II of the β-propeller domain. The newly identified 'site 2' region in Keap1-DC may have a different functional role to regulate Nrf2. Moreover, the crystal structures of Keap1-DC in complex with the DMF analogs, including monoethyl fumarate, fumarate, and itaconate, also exhibited similar binding modes with Keap1-DC. Binding studies confirmed that DMF binds, in a nanomolar range, to the Keap1-DC region as well as the BTB domain of Keap1. Furthermore, the competitive binding assay in the presence of the Nrf2 peptide affirmed the direct binding of DMF at the Nrf2-binding region of Keap1-DC. Overall, our studies suggest that the drug molecule, DMF, binds at multiple sites of Keap1 and thus potentially activates Nrf2 function through covalent as well as the noncovalent mode of action, to combat oxidative stress. DATABASE: Structural data are available in RCSB-protein data bank database(s) under the accession numbers 6LRZ, 7C60, and 7C5E.
Organizational Affiliation:
Department of Biophysics, National Institute of Mental Health and Neuro Sciences (NIMHANS), Bengaluru, India.