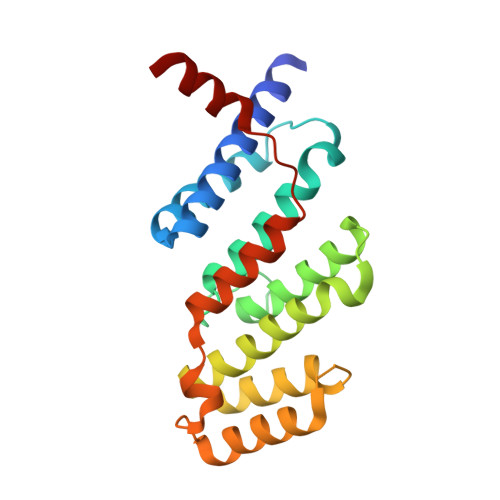
Microcephalin 1/BRIT1-TRF2 interaction promotes telomere replication and repair, linking telomere dysfunction to primary microcephaly.
Cicconi, A., Rai, R., Xiong, X., Broton, C., Al-Hiyasat, A., Hu, C., Dong, S., Sun, W., Garbarino, J., Bindra, R.S., Schildkraut, C., Chen, Y., Chang, S.(2020) Nat Commun 11: 5861-5861
- PubMed: 33203878
- DOI: https://doi.org/10.1038/s41467-020-19674-0
- Primary Citation of Related Structures:
7C5D - PubMed Abstract:
Telomeres protect chromosome ends from inappropriately activating the DNA damage and repair responses. Primary microcephaly is a key clinical feature of several human telomere disorder syndromes, but how microcephaly is linked to dysfunctional telomeres is not known. Here, we show that the microcephalin 1/BRCT-repeats inhibitor of hTERT (MCPH1/BRIT1) protein, mutated in primary microcephaly, specifically interacts with the TRFH domain of the telomere binding protein TRF2. The crystal structure of the MCPH1-TRF2 complex reveals that this interaction is mediated by the MCPH1 330 YRLSP 334 motif. TRF2-dependent recruitment of MCPH1 promotes localization of DNA damage factors and homology directed repair of dysfunctional telomeres lacking POT1-TPP1. Additionally, MCPH1 is involved in the replication stress response, promoting telomere replication fork progression and restart of stalled telomere replication forks. Our work uncovers a previously unrecognized role for MCPH1 in promoting telomere replication, providing evidence that telomere replication defects may contribute to the onset of microcephaly.
Organizational Affiliation:
Department of Laboratory Medicine, Yale University School of Medicine, 330 Cedar St., New Haven, CT, 06520, USA.