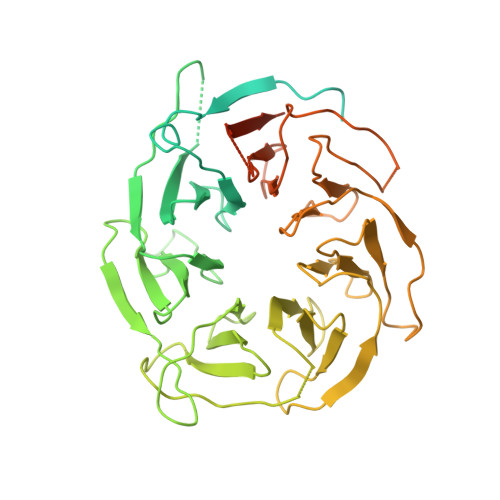
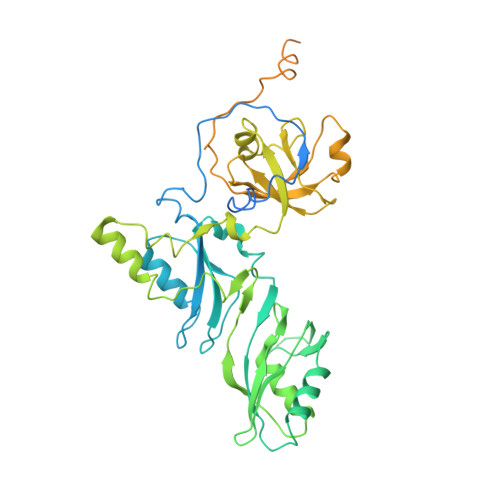
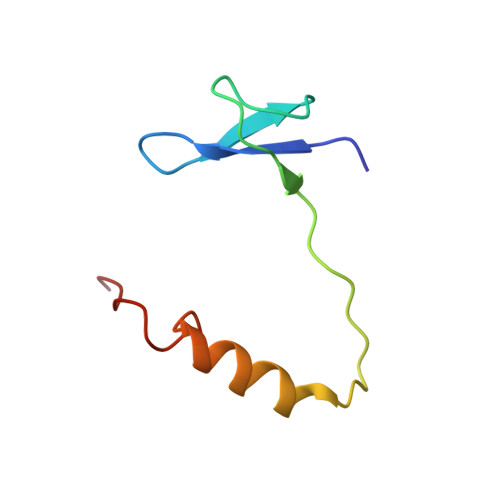
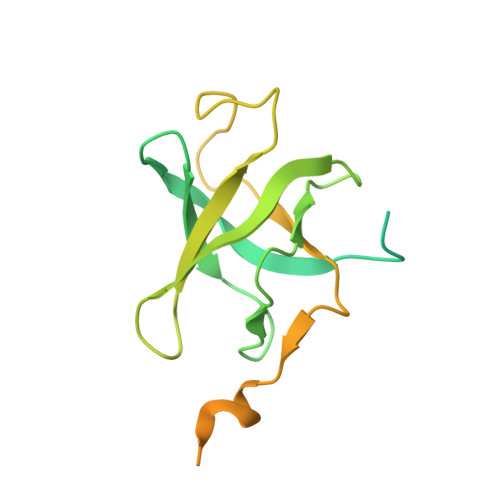
Structure of human telomerase holoenzyme with bound telomeric DNA.
Ghanim, G.E., Fountain, A.J., van Roon, A.M., Rangan, R., Das, R., Collins, K., Nguyen, T.H.D.(2021) Nature 593: 449-453
- PubMed: 33883742
- DOI: https://doi.org/10.1038/s41586-021-03415-4
- Primary Citation of Related Structures:
7BG9, 7BGB - PubMed Abstract:
Telomerase adds telomeric repeats at chromosome ends to compensate for the telomere loss that is caused by incomplete genome end replication 1 . In humans, telomerase is upregulated during embryogenesis and in cancers, and mutations that compromise the function of telomerase result in disease 2 . A previous structure of human telomerase at a resolution of 8 Å revealed a vertebrate-specific composition and architecture 3 , comprising a catalytic core that is flexibly tethered to an H and ACA (hereafter, H/ACA) box ribonucleoprotein (RNP) lobe by telomerase RNA. High-resolution structural information is necessary to develop treatments that can effectively modulate telomerase activity as a therapeutic approach against cancers and disease. Here we used cryo-electron microscopy to determine the structure of human telomerase holoenzyme bound to telomeric DNA at sub-4 Å resolution, which reveals crucial DNA- and RNA-binding interfaces in the active site of telomerase as well as the locations of mutations that alter telomerase activity. We identified a histone H2A-H2B dimer within the holoenzyme that was bound to an essential telomerase RNA motif, which suggests a role for histones in the folding and function of telomerase RNA. Furthermore, this structure of a eukaryotic H/ACA RNP reveals the molecular recognition of conserved RNA and protein motifs, as well as interactions that are crucial for understanding the molecular pathology of many mutations that cause disease. Our findings provide the structural details of the assembly and active site of human telomerase, which paves the way for the development of therapeutic agents that target this enzyme.
- Medical Research Council Laboratory of Molecular Biology, Cambridge, UK.
Organizational Affiliation: