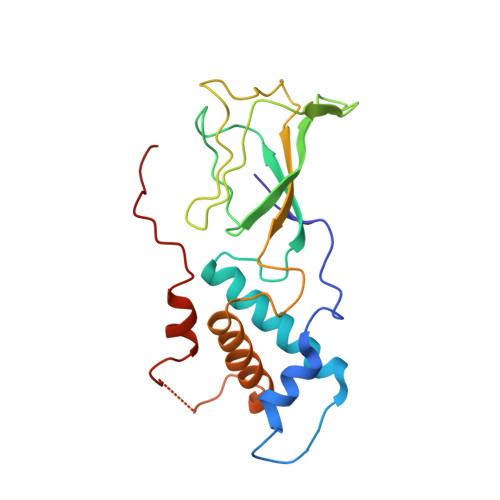
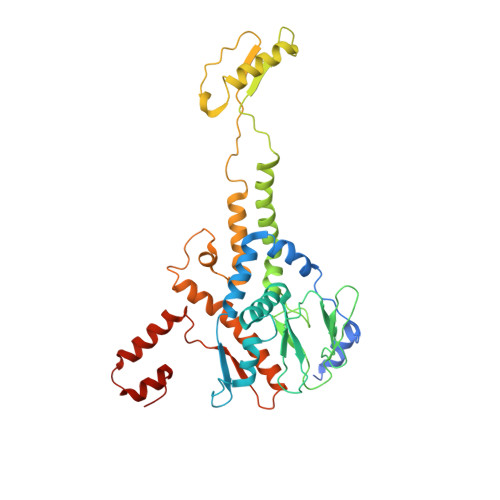
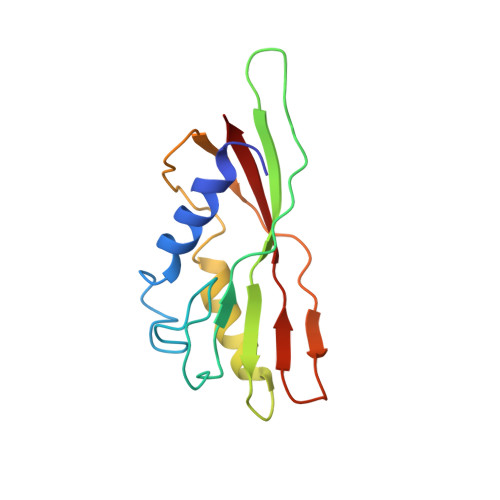
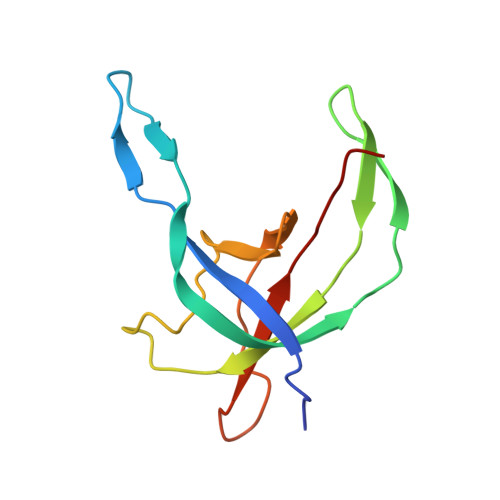
Virion of empty GTA particle
Bardy, P., Fuzik, T., Hrebik, D., Pantucek, R., Beatty, J.T., Plevka, P.(2020) Nat Commun 11: 3034
- PubMed: 32541663
- DOI: https://doi.org/10.1038/s41467-020-16669-9
- Primary Citation of Related Structures:
6TB9, 6TBA, 6TE8, 6TE9, 6TEA, 6TEB, 6TEH, 6TO8, 6TOA, 6TSU, 6TSV, 6TSW, 6TUI - PubMed Abstract:
Alphaproteobacteria, which are the most abundant microorganisms of temperate oceans, produce phage-like particles called gene transfer agents (GTAs) that mediate lateral gene exchange. However, the mechanism by which GTAs deliver DNA into cells is unknown. Here we present the structure of the GTA of Rhodobacter capsulatus (RcGTA) and describe the conformational changes required for its DNA ejection. The structure of RcGTA resembles that of a tailed phage, but it has an oblate head shortened in the direction of the tail axis, which limits its packaging capacity to less than 4,500 base pairs of linear double-stranded DNA. The tail channel of RcGTA contains a trimer of proteins that possess features of both tape measure proteins of long-tailed phages from the family Siphoviridae and tail needle proteins of short-tailed phages from the family Podoviridae. The opening of a constriction within the RcGTA baseplate enables the ejection of DNA into bacterial periplasm.
Organizational Affiliation:
Department of Experimental Biology, Faculty of Science, Masaryk University, 625 00, Brno, Czech Republic.