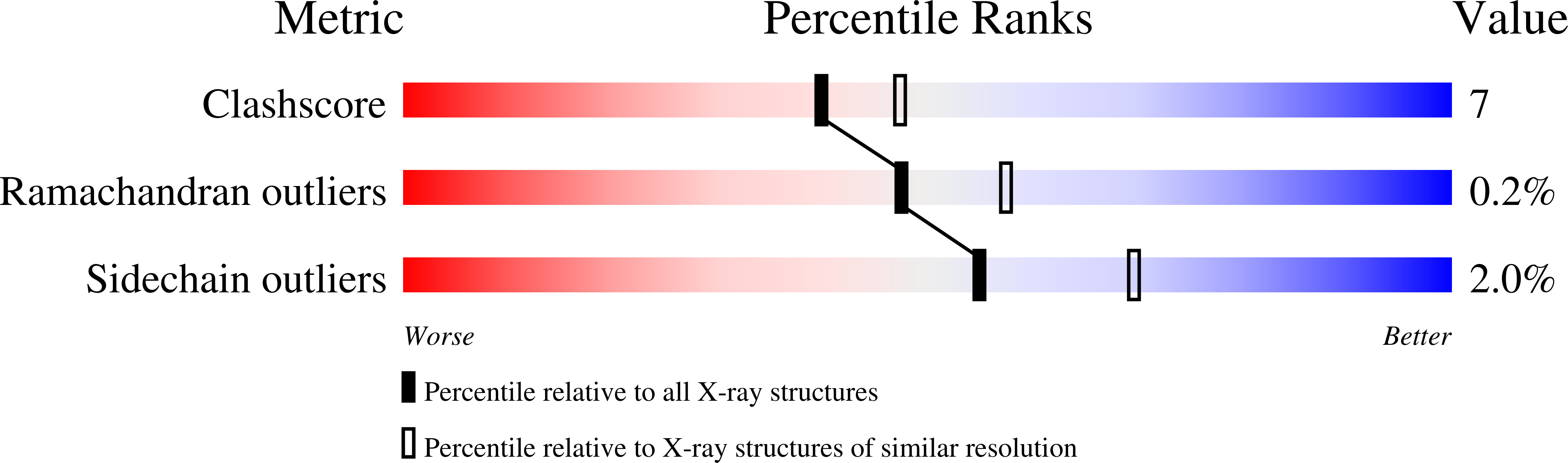A mechanism-inspired UDP- N -acetylglucosamine pyrophosphorylase inhibitor.
Raimi, O.G., Hurtado-Guerrero, R., Borodkin, V., Ferenbach, A., Urbaniak, M.D., Ferguson, M.A.J., van Aalten, D.M.F.(2020) RSC Chem Biol 1: 13-25
- PubMed: 34458745
- DOI: https://doi.org/10.1039/c9cb00017h
- Primary Citation of Related Structures:
6TN3 - PubMed Abstract:
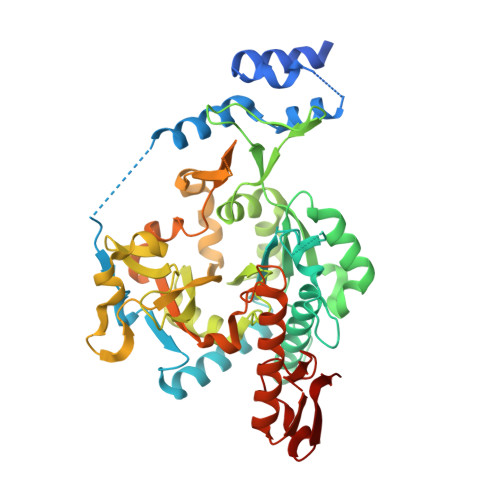
UDP- N -acetylglucosamine pyrophosphorylase (UAP1) catalyses the last step in eukaryotic biosynthesis of uridine diphosphate- N -acetylglucosamine (UDP-GlcNAc), converting UTP and GlcNAc-1P to the sugar nucleotide. Gene disruption studies have shown that this gene is essential in eukaryotes and a possible antifungal target, yet no inhibitors of fungal UAP1 have so far been reported. Here we describe the crystal structures of substrate/product complexes of UAP1 from Aspergillus fumigatus that together provide snapshots of catalysis. A structure with UDP-GlcNAc, pyrophosphate and Mg 2+ provides the first Michaelis complex trapped for this class of enzyme, revealing the structural basis of the previously reported Mg 2+ dependence and direct observation of pyrophosphorolysis. We also show that a highly conserved lysine mimics the role of a second metal observed in structures of bacterial orthologues. A mechanism-inspired UTP α,β-methylenebisphosphonate analogue ( me UTP) was designed and synthesized and was shown to be a micromolar inhibitor of the enzyme. The mechanistic insights and inhibitor described here will facilitate future studies towards the discovery of small molecule inhibitors of this currently unexploited potential antifungal drug target.
Organizational Affiliation:
Division of Gene Regulation and Expression, School of Life Sciences, University of Dundee Dow Street DD1 5EH Dundee UK dmfvanaalten@dundee.ac.uk.