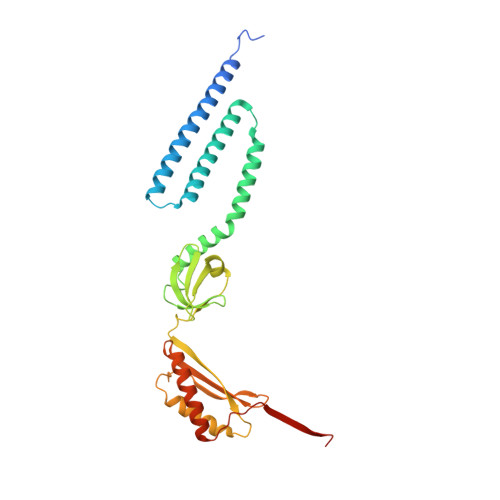
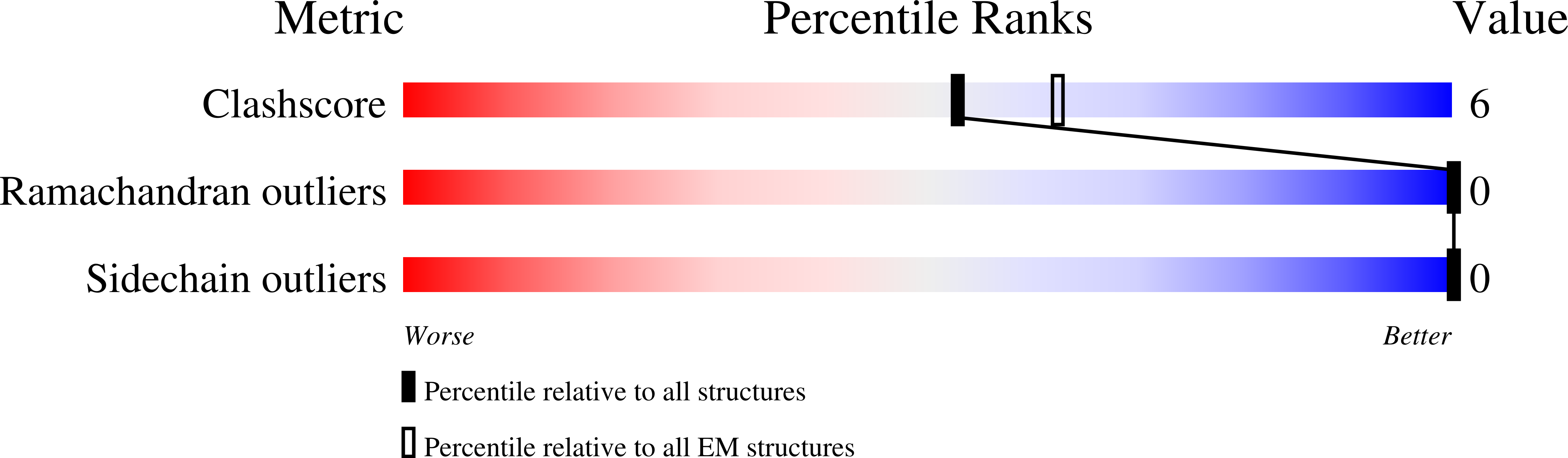Structure of the Mechanosensitive Channel MscS Embedded in the Membrane Bilayer.
Rasmussen, T., Flegler, V.J., Rasmussen, A., Bottcher, B.(2019) J Mol Biol 431: 3081-3090
- PubMed: 31291591
- DOI: https://doi.org/10.1016/j.jmb.2019.07.006
- Primary Citation of Related Structures:
6RLD - PubMed Abstract:
Since life has emerged, gradients of osmolytes over the cell membrane cause pressure changes in the cell and require tight regulation to prevent cell rupture. The mechanosensitive channel of small conductance (MscS) releases solutes and water when a hypo-osmotic shock raises the pressure in the cell. It is a member of a large family of MscS-like channels found in bacteria, archaea, fungi and plants and model for mechanosensation. MscS senses the increase of tension in the membrane directly by the force from the lipids, but the molecular mechanism is still elusive. We determined the lipid interactions of MscS by resolving the structure of Escherichia coli MscS embedded in membrane discs to 2.9-Å resolution using cryo-electron microscopy. The membrane is attached only to parts of the sensor paddles of MscS, but phospholipid molecules move through grooves into remote pockets on the cytosolic side. On the periplasmic side, a lipid bound by R88 at the pore entrance is separated from the membrane by TM1 helices. The N-terminus interacts with the periplasmic membrane surface. We demonstrate that the unique membrane domain of MscS promotes deep penetration of lipid molecules and shows multimodal interaction with the membrane to fine-tune tension sensing.
Organizational Affiliation:
Biocenter and Rudolf Virchow Center, Universität Würzburg, Haus D15, Josef-Schneider-Str. 2, 97080 Würzburg, Germany. Electronic address: tim.rasmussen@uni-wuerzburg.de.