Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6.
Fuchsbauer, O., Swuec, P., Zimberger, C., Amigues, B., Levesque, S., Agudelo, D., Duringer, A., Chaves-Sanjuan, A., Spinelli, S., Rousseau, G.M., Velimirovic, M., Bolognesi, M., Roussel, A., Cambillau, C., Moineau, S., Doyon, Y., Goulet, A.(2019) Mol Cell 76: 922
- PubMed: 31604602
- DOI: https://doi.org/10.1016/j.molcel.2019.09.012
- Primary Citation of Related Structures:
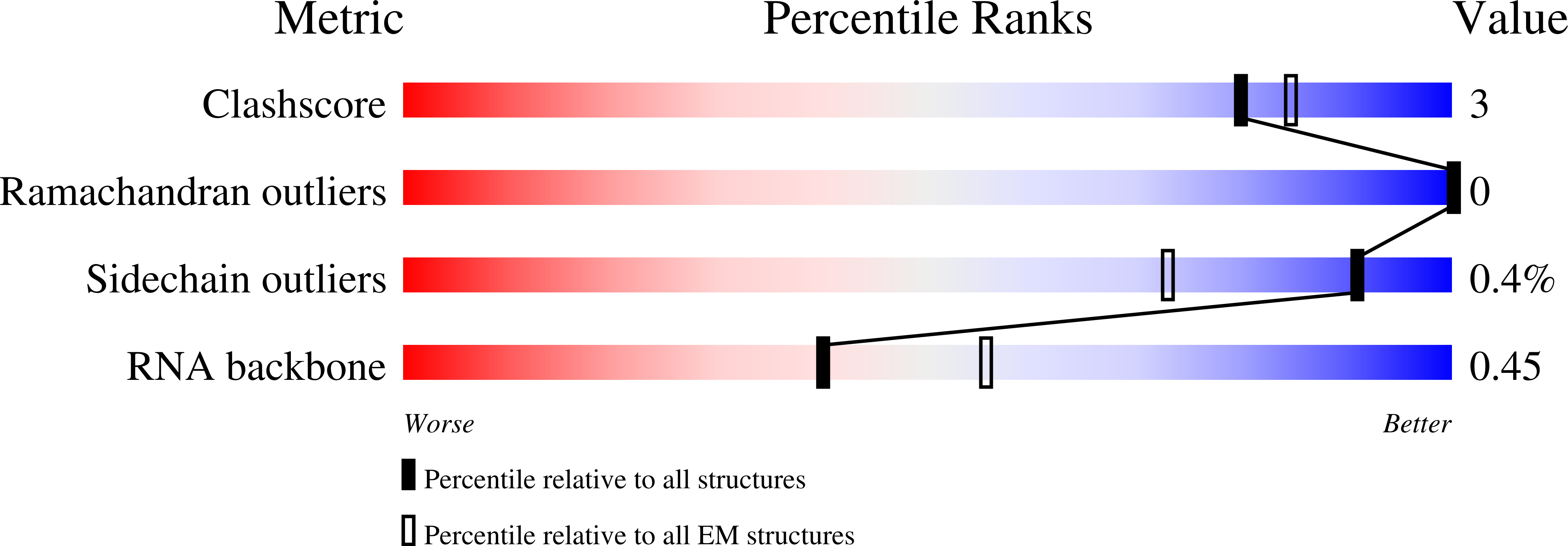
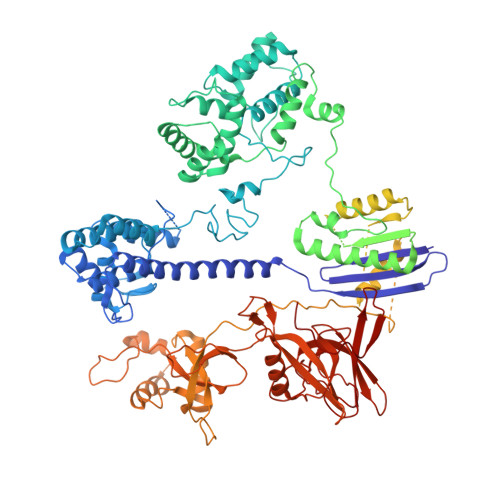
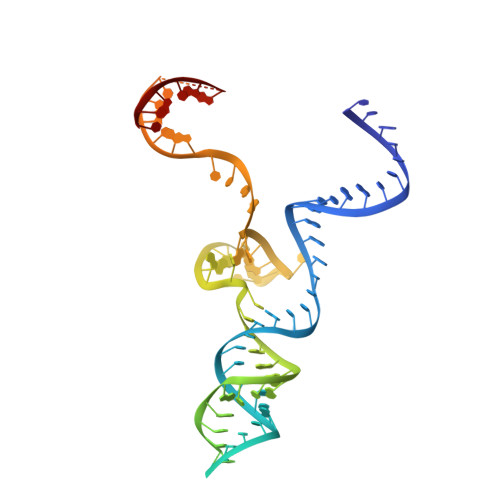
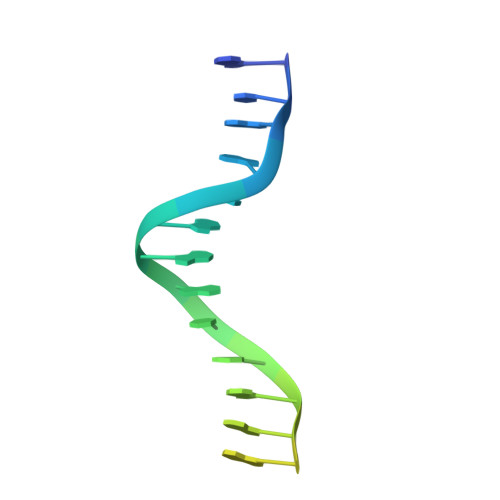
6RJ9, 6RJA, 6RJD, 6RJG - PubMed Abstract:
In the arms race against bacteria, bacteriophages have evolved diverse anti-CRISPR proteins (Acrs) that block CRISPR-Cas immunity. Acrs play key roles in the molecular coevolution of bacteria with their predators, use a variety of mechanisms of action, and provide tools to regulate Cas-based genome manipulation. Here, we present structural and functional analyses of AcrIIA6, an Acr from virulent phages, exploring its unique anti-CRISPR action. Our cryo-EM structures and functional data of AcrIIA6 binding to Streptococcus thermophilus Cas9 (St1Cas9) show that AcrIIA6 acts as an allosteric inhibitor and induces St1Cas9 dimerization. AcrIIA6 reduces St1Cas9 binding affinity for DNA and prevents DNA binding within cells. The PAM and AcrIIA6 recognition sites are structurally close and allosterically linked. Mechanistically, AcrIIA6 affects the St1Cas9 conformational dynamics associated with PAM binding. Finally, we identify a natural St1Cas9 variant resistant to AcrIIA6 illustrating Acr-driven mutational escape and molecular diversification of Cas9 proteins.
Organizational Affiliation:
Architecture et Fonction des Macromolécules Biologiques, Centre National de la Recherche Scientifique (CNRS), Campus de Luminy, Case 932, 13288 Marseille Cedex 09, France; Architecture et Fonction des Macromolécules Biologiques, Aix-Marseille Université, Campus de Luminy, Case 932, 13288 Marseille Cedex 09, France.