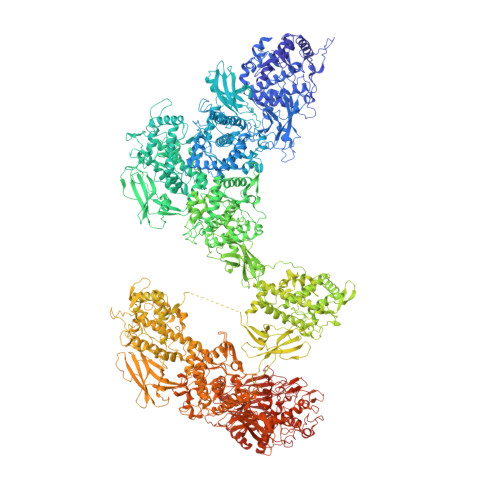
Cryo-EM reveals the asymmetric assembly of squid hemocyanin.
Tanaka, Y., Kato, S., Stabrin, M., Raunser, S., Matsui, T., Gatsogiannis, C.(2019) IUCrJ 6: 426-437
- PubMed: 31098023
- DOI: https://doi.org/10.1107/S205225251900321X
- Primary Citation of Related Structures:
6R83 - PubMed Abstract:
The oxygen transporter of molluscs, hemocyanin, consists of long pearl-necklace-like subunits of several globular domains. The subunits assemble in a complex manner to form cylindrical decamers. Typically, the first six domains of each subunit assemble together to form the cylinder wall, while the C-terminal domains form a collar that fills or caps the cylinder. During evolution, various molluscs have been able to fine-tune their oxygen binding by deleting or adding C-terminal domains and adjusting their inner-collar architecture. However, squids have duplicated one of the wall domains of their subunits instead. Here, using cryo-EM and an optimized refinement protocol implemented in SPHIRE , this work tackled the symmetry-mismatched structure of squid hemocyanin, revealing the precise effect of this duplication on its quaternary structure and providing a potential model for its structural evolution.
Organizational Affiliation:
Graduate School of Life Sciences, Tohoku University, 2-1-1 Katahira, Aoba-ku, Sendai 980-8577, Japan.