Discovery and Optimization of 2-Arylquinazolin-4-ones into a Potent and Selective Tankyrase Inhibitor Modulating Wnt Pathway Activity.
Buchstaller, H.P., Anlauf, U., Dorsch, D., Kuhn, D., Lehmann, M., Leuthner, B., Musil, D., Radtki, D., Ritzert, C., Rohdich, F., Schneider, R., Esdar, C.(2019) J Med Chem 62: 7897-7909
- PubMed: 31381853
- DOI: https://doi.org/10.1021/acs.jmedchem.9b00656
- Primary Citation of Related Structures:
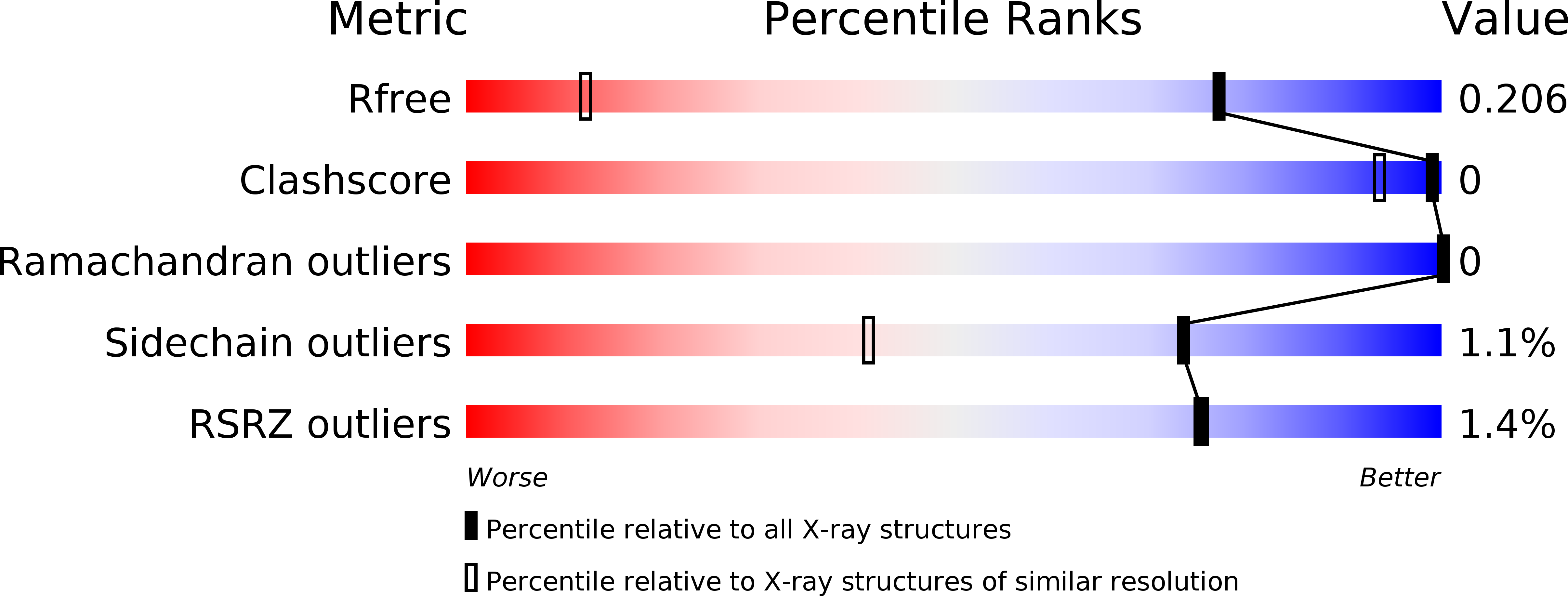
6QXU - PubMed Abstract:
Tankyrases 1 and 2 (TNKS1/2) are promising pharmacological targets that recently gained interest for anticancer therapy in Wnt pathway dependent tumors. 2-Aryl-quinazolinones were identified and optimized into potent tankyrase inhibitors through SAR exploration around the quinazolinone core and the 4'-position of the phenyl residue. These efforts were supported by analysis of TNKS X-ray and WaterMap structures and resulted in compound 5k , a potent, selective tankyrase inhibitor with favorable pharmacokinetic properties. The X-ray structure of 5k in complex with TNKS1 was solved and confirmed the design hypothesis. Modulation of Wnt pathway activity was demonstrated with this compound in a colorectal xenograft model in vivo .
Organizational Affiliation:
Merck Healthcare KGaA , Global Research & Development , Frankfurter Strasse 250 , 64293 Darmstadt , Germany.